Molecular anthropology
[1] This field of anthropology examines evolutionary links between ancient and modern human populations, as well as between contemporary species.
Certain similarities in genetic makeup let molecular anthropologists determine whether or not different groups of people belong to the same haplogroup, and thus if they share a common geographical origin.
This is significant because it allows anthropologists to trace patterns of migration and settlement, which gives helpful insight as to how contemporary populations have formed and progressed over time.
[2] Molecular anthropology has been extremely useful in establishing the evolutionary tree of humans and other primates, including closely related species like chimps and gorillas.
While there are clearly many morphological similarities between humans and chimpanzees, for example, certain studies also have concluded that there is roughly a 98 percent commonality between the DNA of both species.
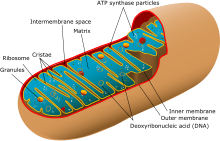
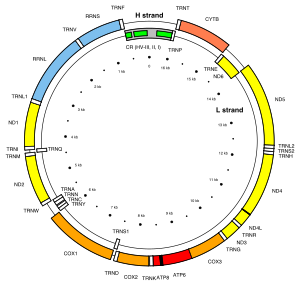
[clarification needed] The non-recombinant portion of the Y chromosome and the mtDNA, under normal circumstances, do not undergo productive recombination.
The process of recombination, if frequent enough, corrupts the ability to create parsimonious trees because of stretches of amino acid subsititions (SNPs).
This allowed the use of mitochondrial DNA to determine that the relative age of the human population was small, having gone through a recent constriction at about 150,000 years ago (see #Causes of errors).
It is unclear whether this is due to high male-to-female ratios in the past coupled with repeat migrations from Africa, as a result of mutational rate change, or as some have even proposed that females of the LCA between chimps and humans continued to pass DNA millions after males ceased to pass DNA.
At present the best evidence suggests that in migration the male to female ratio in humans may have declined, causing a trimming of Y diversity on multiple occasions within and outside of Africa.
One argument that arose was that the Maori by mtDNA appear to have migrated from Eastern China or Taiwan, by Y chromosome from the Papua New Guinea region.
Such admixtures appear to be common in the human population and thus the use of a single haploid loci can give a biased perspective.
The time to most recent common ancestor (TMRCA) ranges from fixed to ~1.8 million years, with a median around 700ky.
These studies roughly plot to the expected fixation distribution of alleles, given linkage disequilibrium between adjacent sites.
There are some distinctions within SSA that suggest a smaller region, but there is not adequate enough sample size and coverage to define a place of most recent common ancestor.
The MCR1 gene has also been sequenced but the results are controversial, with one study claiming that contamination issues cannot be resolved from human Neandertal similarities.
Some of the ancient sequences obtained have highly probable errors, and proper control to avoid contamination.
The molecular phylogenetics is based on quantification substitutions and then comparing sequence with other species, there are several points in the process which create errors.
Unless scientist study the sequence of a great many animals, particularly those close to the branch being examined, they generally do not know what the rate of mutation for a given site.
To date the only way to assess this variance is to apply molecular phylogenetics on species claimed to be branch points.
[6] By 1975 protein sequencing and comparative serology combined were used to propose that humans closest living relative (as a species) was the chimpanzee.
[7] In hindsight, the last common ancestor (LCA) from humans and chimps appears to older than the Sarich and Wilson estimate, but not as old as Leakey claimed, either.
(See Causes of Error) In 1979, W.M.Brown and Wilson began looking at the evolution of mitochondrial DNA in animals, and found they were evolving rapidly.
[8] The technique they used was restriction fragment length polymorphism (RFLP), which was more affordable at the time compared to sequencing.
[16] In 1991 Vigilante et al. published the seminal work on mtDNA phylogeny implicating sub-saharan Africa as the place of humans most recent common ancestors for all mtDNAs.
[17] The war between out-of-Africa and multiregionalism, already simmering with the critiques of Allan Templeton, soon escalated with the paleoanthropologist, like Milford Wolpoff, getting involved.
[18][19][20] In 1995, F. Ayala published his critical Science article "The Myth about Eve", which relied on HLA-DR sequence.
was Harris and Hey, 1998 which showed that the TMCRA (time to most recent common ancestor) for the PDHA1 gene was well in excess of 1 million years.
In 2007, Gonder et al. proposed that a core population of humans, with greatest level of diversity and lowest selection, once lived in the region of Tanzania and proximal parts of southern Africa, since humans left this part of Africa, mitochondria have been selectively evolving to new regions.