Bacterial secretion system
Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors (mainly of proteins) to invade the host cells.
[3] In addition, there is appreciable difference between diderm bacteria with lipopolysaccharide on the outer membrane (diderm-LPS) and those with mycolic acid (diderm-mycolate).
Among Gram-negative bacteria, Escherichia coli, Vibrio cholerae, Klebsiella pneumoniae, and Yersinia enterocolitica use the Sec system.
SRP is a ribonucleoprotein (protein-RNA complex) that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and to the cell membrane in prokaryotes.
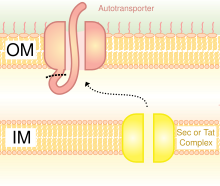
The two pathways require different molecular chaperones and ultimately use a protein-transporting channel SecYEG for transporting the proteins across the inner cell membrane.
[6] In the SecA pathway, SecB acts as a chaperone, helping protein transport to the periplasm after complete synthesis of the peptide chains.
Whereas in the SRP pathway, YidC is the chaperone, and transport proteins to the cell membrane while they are still undergoing peptide synthesis.
[8] However, a recent selective ribosome profiling study suggest that inner membrane proteins with large periplasmic loops are targeted by the SecA pathway.
In SecA pathway, a chaperone trigger factor (TF) first bind to the exposed N-terminal signal sequence of the peptide chain.
[13] Tat proteins are highly variable in different bacteria and are classified into three major types, namely TatA, TatB, and TatC.
[15] Signal peptides that can recognise the Tat proteins are characterised by a consensus motif Ser/Thr-Arg-Arg-X-Phe-Leu-Lys (where X can be any polar amino acid).
[4] It is present in Gram-positive bacteria (as WSS) and Mycobacteria (as Esx in all diderm-mycolates) such as M. tuberculosis, M. bovis, Streptomyces coelicolor and S. aureus.
[21] Type III secretion system (T3SS or TTSS) is structurally similar and related to the basal body of bacterial flagella.
Seen in some of the most virulent Gram-negative bacteria such as Salmonella, Shigella, Yersinia, Vibrio, it is used to inject toxic proteins into eukaryotic cells.
Agrobacterium tumefaciens, from which it was originally discovered, uses this system to send the T-DNA portion of the Ti plasmid into plant cells, in which a crown gall (tumor) is produced as a result.
Legionella pneumophila that causes legionellosis (Legionnaires' disease) has a T4SS called icm/dot (intracellular multiplication/defect in organelle trafficking genes) that transport many bacterial proteins into its eukaryotic host.
[29] Type VI secretion systems (T6SS) were discovered by the team of John Mekalanos at the Harvard Medical School in 2006 from Vibrio cholerae and Pseudomonas aeruginosa.
[32][33] In addition to their classic role as the pathogenicity factor, T6SS are also involved in defense against simple eukaryotic predators and in inter-bacteria interactions.
[4][37] Type IX secretion systems (T9SS) are found regularly in the Fibrobacteres-Chlorobi-Bacteroidetes lineage of bacteria, where member species include an outer membrane.
The system is involved variably in one type of gliding motility, in the proper targeting of certain virulence factors to the cell surface, and the degradation of complex of biopolymers.
At least sixteen structural components of the system have been described, including PorU, a protein-sorting transpeptidase that removes the C-terminal sorting signal from cargo proteins and mediates their attachment instead to lipopolysaccharide.