Bloom filters in bioinformatics
Bloom filters are space-efficient probabilistic data structures used to test whether an element is a part of a set.
This allows the user to manage the risk of a getting a false positive by compromising on the space benefits of the Bloom filter.
BLAST and similar tools cannot handle this problem efficiently, therefore Bloom filter based data structures have been implemented to this end.
BIGSI[6] borrows bitsliced signatures from the field of document retrieval to index and query the entirety of microbial and viral sequencing data in the European Nucleotide Archive.
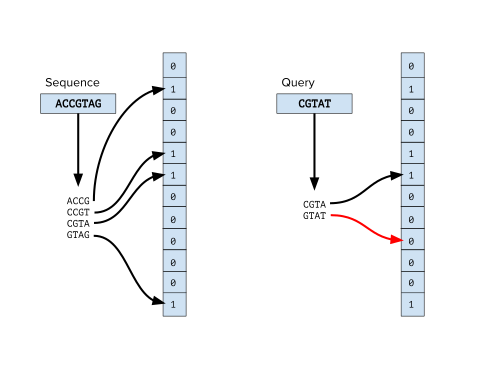
The memory efficiency of Bloom filters has been used in genome assembly as a way to reduce the space footprint of k-mers from sequencing data.
Graph traversal is accomplished by querying the Bloom filter for any of the four possible subsequent k-mers from the current node.
However, these methods have a higher error rate,[10][11] which complicates downstream analysis of the sequence and can even give rise to erroneous conclusions.
Therefore, tools using Bloom filters have been developed to address these limitations, taking advantage of their efficient memory usage.