CAPP-Seq
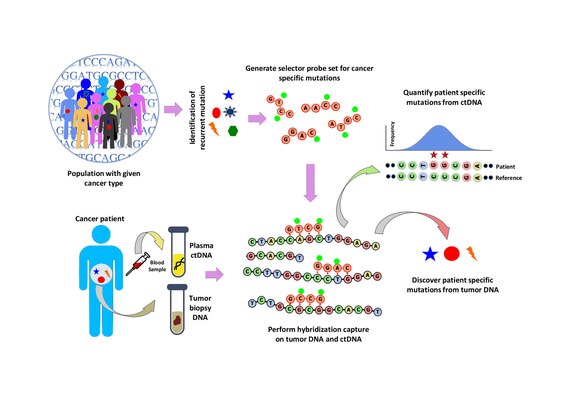
A ‘selector’ is designed which consists of biotinylated DNA oligonucleotide probes targeting the recurrently mutated regions chosen for the specific cancer type.
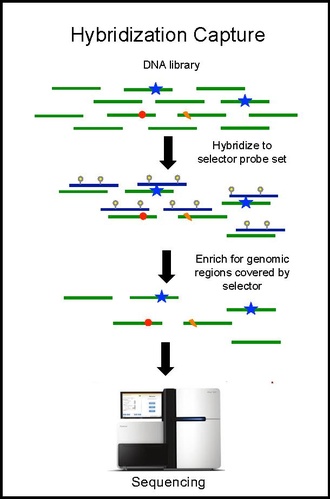
Using the selector, a probe-based hybridization capture is performed on tumor and normal DNA to discover mutations specific to the patient.
In CAPP-Seq, design of selector is a crucial step that identifies recurrent mutations in a particular cancer type using publicly available next generation sequencing data.
Hybridization capture with the selector probe set is performed on tumor DNA from a biopsy and sequenced to a depth of ~10,000× coverage.
The biotinylated selector probes bind selectively to the regions of the DNA library that were chosen to be where the recurrently mutations occur in the given cancer type.
CAPP-Seq can be applied to ctDNA from multiple blood samples at different time points in order to follow tumor evolution.
After the first step of variant calling, germline and loss of heterozygosity (LOH) mutations are removed in CAPP-seq to reduce the background biases.
For example, statistical significance of tumor-derived SNVs can be estimated by random sampling of background alleles using Monte Carlo method.
For the indel calls, statistical significance is calculated applying a separate method that used a strand specific analysis by Z-test shown in previous work.
Sensitivity of this technology depends on the effective design of selector and highly biased with the size of the cohort and type of cancer under study.
The lack of background to find the statistically significant recurrent variants has limited its performance due to stochastic noise and biological variability.
[1] The detection limit of CAPP-Seq is affected by three main areas: the input amount of ctDNA molecules, sample cross-contamination, potential allelic bias in the capture reagent, and PCR or sequencing errors.
Thus, technical sensitivity, reproducibility, specificity and requirement of expertise for analysis of large amount of data are some of the concerned issues with the technique.
[14] Imaging can also be affected by radiation-induced inflammation and fibrotic changes, making it hard to determine if there is residual tumor or just effects of treatment.
In a study [14] in late-stage non-small-cell lung cancer (NSCLC) they found two cases where ctDNA correctly determined the outcome of a patient when medical imaging was wrong.
A simple blood test is non-invasive and much safer and easier to subject cancer patients to multiple times through the course of treatment.
[14] In a study[1] for late stage NSCLC, they performed a version of CAPP-Seq where the tumor biopsy was not sequenced first, and they were able to correctly classify 100% of patient plasma samples with a 0% false positive rate.