Circular RNA
There are three ways in which exon scrambling can occur: The notion that circularized transcripts are byproducts from imperfect splicing is supported by the low abundance and the lack of sequence conservation of most circRNAs,[9] but has been challenged.
[12][13][3][14] It is important that the flanking intronic Alu elements are complementary, as this enables RNA pairing, which in turn facilitates circRNA synthesis.
[16] A-to-I RNA editing in up- and downstream intronic Alu elements flanking the back-splice site (BSS) can reduces the formation of circRNAs in the human heart.
[3] Approximately 1 in 8 expressed genes were found to produce detectable levels of circRNAs, including those of low abundance, which was significantly higher than previously suspected, and was attributed to greater sequencing depth.
IRAlus, either convergent or divergent, are juxtaposed across flanking introns of circRNAs in a parallel way with similar distances to adjacent exons.
[19] Circular RNAs can be separated into five classes:[20][21] A recent study of human circRNAs revealed that these molecules are usually composed of 1–5 exons.
[3] By the Alu repeats base pairing to one another, it has been proposed that this may enable the splice sites to find each other, thus facilitating circularization.
Because the nuclear envelope breaks down during mitosis, one hypothesis is that the molecules exit the nucleus during this phase of the cell cycle.
After digesting total RNA with RNase R, they were able to identify circular species, indicating that circRNAs are not specific to eukaryotes.
For example, both humans and mice encode the HIPK2 and HIPK3 genes, two paralogous kinases which produce a large amount of circRNA from one particular exon in both species.
[30] They directly base-pair to target messenger RNAs (mRNAs), and can trigger cleavage of the mRNA depending on the degree of complementarity.
MicroRNA cleavage activity depends on complementarity beyond the 12th nucleotide position; none of CiRS-7's binding sites meet this requirement.
To silence miR-7 expression in zebrafish, Memczak and colleagues took advantage of a tool called morpholino, which can base pair and sequester target molecules.
[33] Morpholino treatment had the same severe effect on midbrain development as ectopically expressing CiRS-7 in zebrafish brains using injected plasmids.
For example, some areas of the mouse adult hippocampus show expression of CiRS-7 but not miR-7, suggesting that CiRS-7 may have roles that are independent of interacting with the miRNA.
Given its abundance, evolutionary conservation, and potential regulatory role, it is worthwhile to look into how circular RNA can be used to study pathogenesis and devise therapeutic interventions.
For example: Dube et al.,[19] demonstrated for the first time that brain circular RNAs (circRNA) are part of the pathogenic events that lead to Alzheimer's disease, hypothesizing that specific circRNA would be differentially expressed in AD cases compared to controls and that those effects could be detected early in the disease.
A meta-analysis of the discovery and replication results revealed a total of 148 circRNAs that were significantly correlated with CDR after FDR correction.
This is an important study for the field, as it is the first time that circRNA are quantified and validated (by real-time PCR) in human brain samples at genome-wide scale and in large and well-characterized cohorts.
circFOXO3, Titin genes, circSLC8A1-1 and circAmotl1 play an important role in cardiac function through upregulation or inhibition relevant to heart disease.
For example, circAmotl1 overexpression increases cardiomyocyte longevity through binding and translocation of AKT that regulates cardiac repair.
Although the relevance of circular RNA overexpression and downregulation to heart disease has been found from various research studies, it is still unclear.
Therefore, further research is needed to trace disease progression in different stages of cardiac dysfunction using circular RNA as a biomarker and can be used for gene delivery purposes in cells.
In these circumstances circular RNA proves to be a novel biomarker and is also used for targeted therapy of kidney disease because its pseudogene can alter DNA composition.
[44][45] Circular RNA has a function in autoimmune disease progression acting as a miRNA sponge which regulates DNA methylation, adaptive immune activation, and costimulatory molecule secretion.
Treatment with circular RNA activates the differentiation and maturation of dendritic cells which then secrete a large number of different cytokines and chemokines by expressing the genes for IL-1β, IL-6 and TNFa.
Circular RNA has the very advantageous properties of stability and long shelf life, so it is useful for use as biomarkers and plasmids to express genes of interest.
[47] Viroids are mostly plant pathogens, which consist of short stretches (a few hundred nucleobases) of highly complementary, circular, single-stranded, and non-coding RNAs without a protein coat.
Compared with other infectious plant pathogens, viroids are extremely small in size, ranging from 246 to 467 nucleobases; they thus consist of fewer than 10,000 atoms.
[48] It has been proposed that circular RNA genomes which are common in the modern living world may have had their origin at the beginning of life.
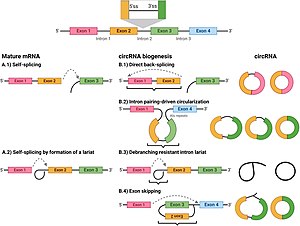
B.1) direct back splicing;
B.2) Intron pairing driven circularization;
B.3) debranching resistant intron lariat ;
B.4) lariat-driven circularization ( exon skipping ).