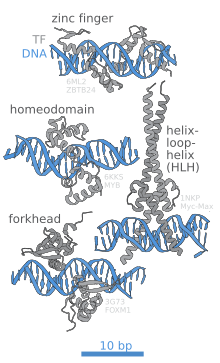
DNA-binding protein
[3][4][5] Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair.
DNA-binding proteins can incorporate such domains as the zinc finger, the helix-turn-helix, and the leucine zipper (among many others) that facilitate binding to nucleic acid.
[8][9] The histones form a disk-shaped complex called a nucleosome, which contains two complete turns of double-stranded DNA wrapped around its surface.
These non-specific interactions are formed through basic residues in the histones making ionic bonds to the acidic sugar-phosphate backbone of the DNA, and are therefore largely independent of the base sequence.
[13] Biophysical studies show that these architectural HMG proteins bind, bend and loop DNA to perform its biological functions.
[21] Thus, these proteins are often the targets of the signal transduction processes that control responses to environmental changes or cellular differentiation and development.
The following lists some methods currently in use:[29] Electrophoretic mobility shift assay (EMSA) is a widespread qualitative technique to study protein–DNA interactions of known DNA binding proteins.
[30][31] DNA-Protein-Interaction - Enzyme-Linked ImmunoSorbant Assay (DPI-ELISA) allows the qualitative and quantitative analysis of DNA-binding preferences of known proteins in vitro.
[32][33] This technique allows the analysis of protein complexes that bind to DNA (DPI-Recruitment-ELISA) or is suited for automated screening of several nucleotide probes due to its standard ELISA plate formate.
[34] [35] DNase footprinting assay can be used to identify the specific sites of binding of a protein to DNA at basepair resolution.
The protein–DNA interactions can be modulated using stimuli like ionic strength of the buffer, macromolecular crowding,[27] temperature, pH and electric field.