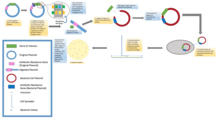
Molecular cloning
The recombinant DNA is then introduced into a host organism (typically an easy-to-grow, benign, laboratory strain of E. coli bacteria).
This single cell can then be expanded exponentially to generate a large number of bacteria, each of which contains copies of the original recombinant molecule.
[5] Prior to the 1970s, the understanding of genetics and molecular biology was severely hampered by an inability to isolate and study individual genes from complex organisms.
Although the detailed planning of the cloning can be done in any text editor, together with online utilities for e.g. PCR primer design, dedicated software exist for the purpose.
Notably, the growing capacity and fidelity of DNA synthesis platforms allows for increasingly intricate designs in molecular engineering.
These shifts introduce complexity that require design to move away from the flat nucleotide-based representation and towards a higher level of abstraction.
E. coli and plasmid vectors are in common use because they are technically sophisticated, versatile, widely available, and offer rapid growth of recombinant organisms with minimal equipment.
The DNA is then purified using simple methods to remove contaminating proteins (extraction with phenol), RNA (ribonuclease) and smaller molecules (precipitation and/or chromatography).
Polymerase chain reaction (PCR) methods are often used for amplification of specific DNA or RNA (RT-PCR) sequences prior to molecular cloning.
Such a designed sequence may be required when moving genes across genetic codes (for example, from the mitochondria to the nucleus)[14] or simply for increasing expression via codon optimization.
[15] The purified DNA is then treated with a restriction enzyme to generate fragments with ends capable of being linked to those of the vector.
If necessary, short double-stranded segments of DNA (linkers) containing desired restriction sites may be added to create end structures that are compatible with the vector.
[3][12] The DNA mixture, previously manipulated in vitro, is moved back into a living cell, referred to as the host organism.
In these vectors, foreign DNA is inserted into a sequence that encodes an essential part of beta-galactosidase, an enzyme whose activity results in formation of a blue-colored colony on the culture medium that is used for this work.
This may be accomplished through a very wide range of experimental methods, including the use of nucleic acid hybridizations, antibody probes, polymerase chain reaction, restriction fragment analysis and/or DNA sequencing.
[3][12] Molecular cloning provides scientists with an essentially unlimited quantity of any individual DNA segments derived from any genome.
In practice, it is frequently more difficult to develop an organism that produces an active form of the recombinant protein in desirable quantities than it is to clone the gene.
This is because the molecular signals for gene expression are complex and variable, and because protein folding, stability and transport can be very challenging.
Although most GMOs are generated for purposes of basic biological research (see for example, transgenic mouse), a number of GMOs have been developed for commercial use, ranging from animals and plants that produce pharmaceuticals or other compounds (pharming), herbicide-resistant crop plants, and fluorescent tropical fish (GloFish) for home entertainment.
The first is alteration of germ cells, that is, sperm or eggs, which results in a permanent genetic change for the whole organism and subsequent generations.
Clinical trials of somatic cell gene therapy began in the late 1990s, mostly for the treatment of cancers and blood, liver, and lung disorders.
[23] Despite a great deal of publicity and promises, the history of human gene therapy has been characterized by relatively limited success.
[23] The effect of introducing a gene into cells often promotes only partial and/or transient relief from the symptoms of the disease being treated.