DNA polymerase II
[7] As characterized, this new mutant strain was more sensitive to ultraviolet light, corroborating the hypothesis that DNA pol I was involved in repair replication.
The mutant grew at the same rate as the wild type, indicating the presence of another enzyme responsible for DNA replication.
The isolation and characterization of this new polymerase involved in semiconservative DNA replication followed, in parallel studies conducted by several labs.
[10] DNA Pol II is an 89.9 kD protein, composed of 783 amino acids, that is encoded by the polB (dinA) gene.
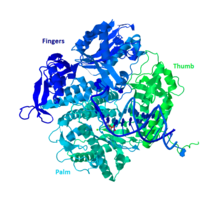
A globular protein, DNA Pol II functions as a monomer, whereas many other polymerases will form complexes.
The palm of the complex contains three catalytic residues that will coordinate with two divalent metal ions in order to function.
The other members of group B do have at least one other subunit which makes the DNA Pol II unique.
[11] Polymerases all are involved with DNA replication in some capacity, synthesizing chains of nucleic acids.
While DNA Pol II is not a major factor in chromosome replication, it has other roles to fill.
While it might not be as fast as DNA Pol III, it has some abilities that make it an effective enzyme.
One important role that DNA Pol II is the major polymerase for the repairing of inter-strand cross-links.
Interstrand cross-links are caused by chemicals such as nitrogen mustard and psoralen which create cytotoxic lesions.
The exact mechanism of how these interstrand cross-links are fixed is still being researched, but it is known that Pol II is highly involved.
[12] In order to fix an error in the sequence, DNA Pol II catalyzes the repair of nucleotide base pairs.
The correct dNTP is then bound and the enzyme complex undergoes conformational changes of subdomains and amino acid residues.