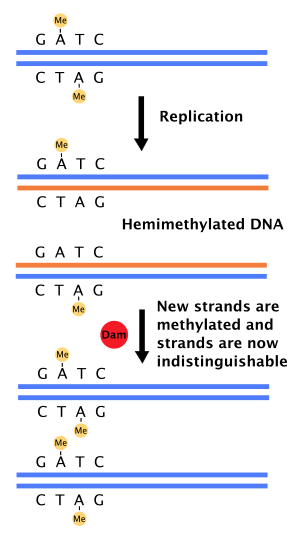
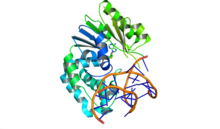
DNA adenine methylase
[4] A repair enzyme, MutS, binds to mismatches in DNA and recruits MutL, which subsequently activates the endonuclease MutH.
Part of this can be explained by the slow hydrolysis of ATP by DnaA, a protein that binds to repeats in the oriC to initiate replication.
[16] A knockout of the dam gene in Aggregatibacter actinomycetemcomitans resulted in dysregulated levels of the protein, leukotoxin, and also reduced the microbe's ability to invade oral epithelial cells.
[15] Additionally a study on Dam methylase deficient Streptococcus mutans, a dental pathogen, revealed the dysregulation of a 103 genes some of which include cariogenic potential.
[17] Motif I consists of a Gly-X-Gly tripeptide and is referred to as the G-loop and is implicated in the binding of the S-Adenosyl methionine cofactor.
[18] Motif II is highly conserved among N4 and N6-adenine methylases and contains a negatively charged amino acid followed by a hydrophobic side chain in the last positions of the β2 strand to bind the AdoMet.
[17] A 2015 crystallography experiment showed that E. coli Dam methylase was able to bind non-GATC DNA with the same sequence of motifs discussed; the authors posit that the obtained structure could serve as grounds for repression of transcription that is not methylation based.
[20] Dam methylase is an orphan methyltransferase that is not part of a restriction-modification system but operates independently to regulate gene expression, mismatch repair, and bacterial replication amongst many other functions.