Ubiquitin ligase
The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein.
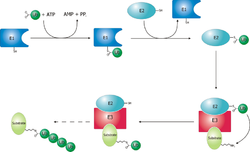
In the conserved first step, an E1 cysteine residue attacks the ATP-activated C-terminal glycine on ubiquitin, resulting in a thioester Ub-S-E1 complex.
[5] The final step in the first ubiquitylation event is an attack from the target protein lysine amine group, which will remove the cysteine, and form a stable isopeptide bond.
For example, phosphorylation of the Tyrosine at position 1045 in the Epidermal Growth Factor Receptor (EGFR) can recruit the RING type E3 ligase c-Cbl, via an SH2 domain.
This can be achieved by different mechanisms, most of which involve recognition of degrons: specific short amino acid sequences or chemical motifs on the substrate.
According to the N-end rule, different N-terminal amino acids (or N-degrons) are recognized to a different extent by their appropriate ubiquitin ligase (N-recognin), influencing the half-life of the protein.
[16] For instance, positively charged (Arg, Lys, His) and bulky hydrophobic amino acids (Phe, Trp, Tyr, Leu, Ile) are recognized preferentially and thus considered destabilizing degrons since they allow faster degradation of their proteins.
[18] The von Hippel-Lindau (VHL) protein (substrate recognition part of a specific E3 ligase), for instance, recognizes the hypoxia-inducible factor alpha (HIF-α) only under normal oxygen conditions, when its proline is hydroxylated.
In addition to recognizing amino acids, ubiquitin ligases can also detect unusual features on substrates that serve as signals for their destruction.
[14] Misfolded or excess unassembled glycoproteins of the ERAD pathway, on the other hand, are recognized by Fbs1 and Fbs2, mammalian F-box proteins of E3 ligases SCFFbs1and SCFFbs2.
This relation can be demonstrated with TRF1 protein (regulator of human telomere length), which is recognized by its corresponding E3 ligase (FBXO4) via an intermolecular beta sheet interaction.
[14] E3 ubiquitin ligases regulate homeostasis, cell cycle, and DNA repair pathways, and as a result, a number of these proteins are involved in a variety of cancers, including famously MDM2, BRCA1, and Von Hippel-Lindau tumor suppressor.