Ribosomal frameshift
In December 2023, it was reported that in vitro-transcribed (IVT) mRNAs in response to BNT162b2 (Pfizer–BioNTech) anti-COVID-19 vaccine caused ribosomal frameshifting.
In eukaryotes it appears to play a role in regulating gene expression levels by generating premature stops and producing nonfunctional transcripts.
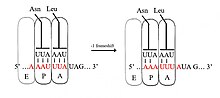
There are typically three elements that comprise a −1 frameshift signal: a slippery sequence, a spacer region, and an RNA secondary structure.
The slippery sequence fits a X_XXY_YYH motif, where XXX is any three identical nucleotides (though some exceptions occur), YYY typically represents UUU or AAA, and H is A, C or U.
[2][11] In this model, the motif structure is explained by the fact that the first and second positions of the anticodons must be able to pair perfectly in both the 0 and −1 frames.
[14] Due to this lag, there exist in small sections of codons sequences that control the rate of ribosomal frameshifting.
In programmed −1 ribosomal frameshifting, the slippery sequence fits a X_XXY_YYH motif, where XXX is any three identical nucleotides (though some exceptions occur), YYY typically represents UUU or AAA, and H is A, C or U.
[16] Efficient ribosomal frameshifting generally requires the presence of an RNA secondary structure to enhance the effects of the slippery sequence.
[12] The RNA structure (which can be a stem-loop or pseudoknot) is thought to pause the ribosome on the slippery site during translation, forcing it to relocate and continue replication from the −1 position.
[2] This model is supported by the fact that strength of the pseudoknot has been positively correlated with the level of frameshifting for associated mRNA.
The rules of the International Union of Pure and Applied Chemistry (IUPAC) are as follows:[18] These symbols are also valid for RNA, except with U (uracil) replacing T (thymine).
Certain proteins which are needed for codon recognition or which bind directly to the mRNA sequence have also been shown to modulate frameshifting levels.