Ribosomal pause
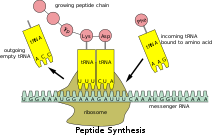
These transcripts are decoded and converted into an amino acid sequence during protein synthesis by ribosomes.
The main reason for these differences was thought to be the concentration of varieties of rare tRNAs limiting the rate at which some transcripts could be decoded.
[2] Two techniques can localize the ribosomal pause site in vivo; a micrococcal nuclease protection assay and isolation of polysomal transcript.
[6] Isolation of polysomal transcripts occurs by centrifuging tissue extracts through a sucrose cushion with translation elongation inhibitors, for example cycloheximide.
[8] Some of the elongation inhibitors, such as: cycloheximide (in eukaryotes) or chloramphenicol, cause the ribosomes to pause and to accumulate in the start codons.
[9] Some forms of ribosomal pause are reversible without needing to discard the translated peptide and mRNA.
[11] More severe "stalls" can be caused an actual lack of tRNA or by the mRNA terminating without a stop codon.
[4] In this case, ribosomal quality control (RQC) performs crisis rescue by translational abandonment.
The incomplete polypeptide is targeted for destruction; in eukaryotes, mRNA no-go decay is also triggered.
The ribosome pause position will help to identify the mRNA sequence features, structure, and the transacting factor that modulates this process.
When the kinetics layer is added,[18] it discloses the time of the pause, and the translation takes place.