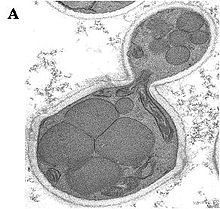
Ogataea polymorpha
The synthetic steps for trehalose synthesis have been detailed for O. polymorpha, and TPS1, the key enzyme gene of this pathway, has been isolated and characterized.
Growth on methanol is accompanied by a massive proliferation of cell organelles named peroxisomes in which the initial enzymatic steps of this pathway take place.
An especially high abundance can be observed for enzymes called MOX (methanol oxidase), FMDH (formate dehydrogenase), and DHAS (dihydroxyacetone synthase).
O. polymorpha produces glycoproteins with two types of sugar chains, N- and O-linked glycans are attached to protein.
[citation needed] Ogataea polymorpha with its unusual characteristics provides an excellent platform for the gene technological production of proteins, especially of pharmaceuticals like insulin for treatment of diabetes, hepatitis B vaccines or IFNalpha-2a for the treatment of hepatitis C. Derivatives of both CBS4732 and DL-1 are employed in the production of such recombinant compounds.
Like other yeasts, O. polymorpha is a microorganism that can be cultured in large fermenters to high cell densities within a short time.
It can release compounds into a culture medium as it has all the components required for secretion (this is for instance not the case with bacteria like Escherichia coli).
This means that the segment following the promoter is variable depending on the desired product – it could be a sequence determining the amino acids for insulin, for hepatitis B vaccine or for interferon.
The attractiveness of the O. polymorpha platform is commercially exploited by several biotech companies for the development of production processes, among others by PharmedArtis, located in Aachen, Germany and the Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK).