Hydrogenase
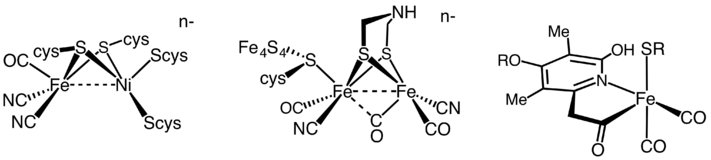
[NiFe] and [FeFe] hydrogenases have some common features in their structures: Each enzyme has an active site and a few Fe-S clusters that are buried in protein.
Another [NiFe], called Huc or Hyd1 or cyanobacterial-type uptake hydrogenase,[10] has been found to be oxygen insensitive while having a very high affinity for hydrogen.
The H-cluster consists of a [4Fe4S] cubane-shaped structure, coupled to the low valent diiron co-factor by a cysteine derived thiol.
[18] In order to conserve energy, anaerobic bacteria use electron bifurcation where exergonic and endergonic redox reactions are coupled to circumvent thermodynamic barriers.
[25] 5,10-methenyltetrahydromethanopterin hydrogenase (EC 1.12.98.2) found in methanogenic Archaea contains neither nickel nor iron-sulfur clusters but an iron-containing cofactor that was recently characterized by X-ray diffraction.
In [NiFe] and [FeFe] hydrogenases, electrons travel through a series of metallorganic clusters that comprise a long distance; the active site structures remain unchanged during the whole process.
While the exact mechanism of the catalysis is still under study, recent finding suggests that molecular hydrogen is first heterolytically cleaved by Fe(II), followed by transfer of hydride to the carbocation of the acceptor.
One popular approach employs mutagenesis to elucidate roles of amino acids and/or ligands in different steps of catalysis such as intramolecular transport of substrates.
For instance, Cornish et al. conducted mutagenesis studies and found out that four amino acids located along the putative channel connecting the active site and protein surface are critical to enzymatic function of [FeFe] hydrogenase from Clostridium pasteurianum (CpI).
In fact, Cao and Hall combined both approaches in developing the model that describes how hydrogen molecules are oxidized or produced within the active site of [FeFe] hydrogenases.
[31] While more research and experimental data are required to complete our understanding of the mechanism, these findings have allowed scientists to apply the knowledge in, e.g., building artificial catalysts mimicking active sites of hydrogenases.
[32] Assuming that the Earth's atmosphere was initially rich in hydrogen, scientists hypothesize that hydrogenases were evolved to generate energy from/as molecular H2.
[33] Microbial communities driven by molecular hydrogen have, in fact, been found in deep-sea settings where other sources of energy from photosynthesis are not available.
The soluble [NiFe] hydrogenase from Ralstonia eutropha H16 is a promising candidate enzyme for H2-based biofuel application as it favours H2 oxidation and is relatively oxygen-tolerant.
[9] Understanding the catalytic mechanism of hydrogenase might help scientists design clean biological energy sources, such as algae, that produce hydrogen.
[5][39] For instance, Stripp et al. relied on protein film electrochemistry and discovered that O2 first converts into a reactive species at the active site of [FeFe] hydrogenases, and then damages its [4Fe-4S] domain.
Bingham et al.'s recent success in engineering [FeFe] hydrogenase from Clostridium pasteurianum was also limited to retained activity (during exposure to oxygen) for H2 consumption only.
[9][4][43] The bidirectional or reversible reaction catalyzed by hydrogenase allows for the capture and storage of renewable energy as fuel with use on demand.
This can be demonstrated through the chemical storage of electricity obtained from a renewable source (e.g. solar, wind, hydrothermal) as H2 during periods of low energy demands.