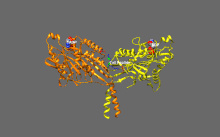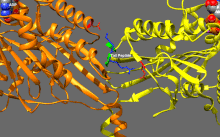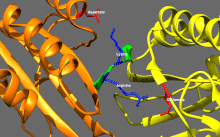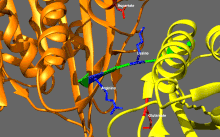Kinesin
The first kinesins to be discovered were microtubule-based anterograde intracellular transport motors[2] in 1985, based on their motility in cytoplasm extruded from the giant axon of the squid.
[3] The founding member of this superfamily, kinesin-1, was isolated as a heterotetrameric fast axonal organelle transport motor consisting of four parts: two identical motor subunits (called Kinesin Heavy Chain (KHC) molecules) and two other molecules each known as a Kinesin Light Chain (KLC).
[6] Molecular genetic and genomic approaches have led to the recognition that the kinesins form a diverse superfamily of motors that are responsible for multiple intracellular motility events in eukaryotic cells.
ATP binding and hydrolysis as well as ADP release change the conformation of the microtubule-binding domains and the orientation of the neck linker with respect to the head; this results in the motion of the kinesin.
Kinesins are motor proteins that transport such cargo by walking unidirectionally along microtubule tracks hydrolysing one molecule of adenosine triphosphate (ATP) at each step.
[21] It was thought that ATP hydrolysis powered each step, the energy released propelling the head forwards to the next binding site.
[30] However, it has been recently discovered that in budding yeast cells kinesin Cin8 (a member of the Kinesin-5 family) can move toward the minus end as well, or retrograde transport.
[31][32][33] Kinesin, so far, has only been shown to move toward the minus end when in a group, with motors sliding in the antiparallel direction in an attempt to separate microtubules.
One specific study tested the speed at which Cin8 motors moved, their results yielded a range of about 25-55 nm/s, in the direction of the spindle poles.
[36] This discovery in kinesin-14 family proteins (such as Drosophila melanogaster NCD, budding yeast KAR3, and Arabidopsis thaliana ATK5) allows kinesin to walk in the opposite direction, toward microtubule minus end.
There is a need especially for approaches which better make a link with the molecular architecture of the protein and data obtained from experimental investigations.
Seiferth et al. demonstrated how quantities such as the velocity or the entropy production of a motor change when adjacent states are merged in a multi-cyclic model until eventually the number of cycles is reduced.
The normal totally asymmetric simple exclusion process for (or TASEP) results can be recovered from this model making the energy equal to zero.
In recent years, it has been found that microtubule-based molecular motors (including a number of kinesins) have a role in mitosis (cell division).