Methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features.
p53 is a known tumor suppressor that activates DNA repair pathways, initiates apoptosis, and pauses the cell cycle.
[3] Natural product methyltransferases provide a variety of inputs into metabolic pathways, including the availability of cofactors, signalling molecules, and metabolites.
This increases the strength of the positive charge and residue hydrophobicity, allowing other proteins to recognize methyl marks.
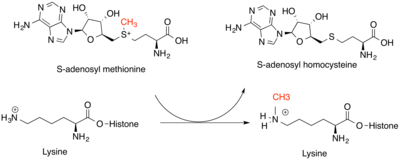
[4] Methyl marks on the histones contribute to these changes by serving as sites for recruitment of other proteins that can further modify chromatin.
[5] N-alpha methyltransferases transfer a methyl group from SAM to the N-terminal nitrogen on protein targets.
DNA methylation, a key component of genetic regulation, occurs primarily at the 5-carbon of the base cytosine, forming 5’methylcytosine (see left).
These enzymes use S-adenosylmethionine as a methyl donor and contain several highly conserved structural features between the three forms; these include the S-adenosylmethionine binding site, a vicinal proline-cysteine pair which forms a thiolate anion important for the reaction mechanism, and the cytosine substrate binding pocket.
In addition to controlling the expression of certain genes, there are a variety of protein complexes, many with implications for human health, which only bind to methylated DNA recognition sites.
m6A methyltransferases methylate the amino group in DNA at C-6 position specifically to prevent the host system to digest own genome through restriction enzymes.
The methylated products of these reactions serve a variety of functions, including co-factors, pigments, signalling compounds, and metabolites.
[15] Catechol-O-methyltransferase (COMT) degrades a class of molecules known as catecholamines that includes dopamine, epinephrine, and norepenepherine.
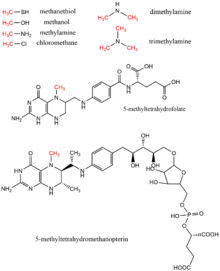
[17] These substrates contribute to methyl transfer pathways including methionine biosynthesis, methanogenesis, and acetogenesis.
Based on different protein structures and mechanisms of catalysis, there are 3 different types of radical SAM (RS) methylases: Class A, B, and C. Class A RS methylases are the best characterized of the 4 enzymes and are related to both RlmN and Cfr.
Plasmid vectors capable of transmitting this gene are a cause of potentially dangerous cross resistance.
[20] Examples of methyltransferase enzymes relevant to disease: Recent work has revealed the methyltransferases involved in methylation of naturally occurring anticancer agents to use S-Adenosyl methionine (SAM) analogs that carry alternative alkyl groups as a replacement for methyl.
[22] The role and potential application of m5C includes to balance the impaired DNA in cancer both hypermethylation and hypomethylation.