MicroRNA sequencing
Evidence that dysregulated miRNAs play a role in diseases such as cancer[1] has positioned miRNA-seq to potentially become an important tool in the future for diagnostics and prognostics as costs continue to decrease.
[2] Like other miRNA profiling technologies, miRNA-Seq has both advantages (sequence-independence, coverage) and disadvantages (high cost, infrastructure requirements, run length, and potential artifacts).
[4][5][6] The first miRNA to be discovered, lin-4, was found in a genetic mutagenesis screen to identify molecular elements controlling post-embryonic development of the nematode Caenorhabditis elegans.
[7] The lin-4 gene encoded a 22 nucleotide RNA with conserved complementary binding sites in the 3’-untranslated region of the lin-14 mRNA transcript[8] and downregulated LIN-14 protein expression.
[9] miRNAs are now thought to be involved in the regulation of many developmental and biological processes, including haematopoiesis (miR-181 in Mus musculus[10]), lipid metabolism (miR-14 in Drosophila melanogaster[11]) and neuronal development (lsy-6 in Caenorhabditis elegans[12]).
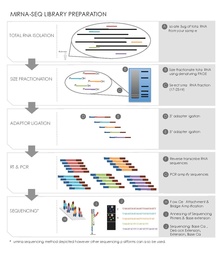
Sequencing preparation involved creating libraries by cloning of DNA reverse transcribed from endogenous small RNAs of 21–25 bp size selected by column and gel electrophoresis.
[17] Applied Biosystems SOLiD sequencing platform has also been used to examine the prognostic value of miRNAs in detecting human breast cancer.
[19][20] Reverse Transcription and PCR Amplification This step converts the small adaptor ligated RNAs into cDNA clones used in the sequencing reaction.
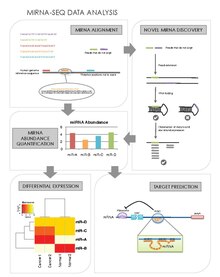
[21][26] Their general steps are as follows:[27] Another advantage of miRNA-seq is that it allows the discovery of novel miRNAs that may have eluded traditional screening and profiling methods.
[41][42] Validation of target cleavage in specific mRNAs is typically performed using a modified version of 5' Rapid Amplification of cDNA Ends with a gene-specific primer.
Consequently, it is not unexpected that miRNAs are involved in various aspects of cancer through the regulation of onco- and tumor suppressor gene expression.
In combination with the development of high-throughput profiling methods, miRNAs have been identified as biomarkers for cancer classification, response to therapy, and prognosis.
[48] Additionally, because miRNAs regulate gene expression they can also reveal perturbations in important regulatory networks that may be driving a particular disorder.