Pyrosequencing
The principle of pyrosequencing was first described in 1993[1] by, Bertil Pettersson, Mathias Uhlen and Pål Nyren by combining the solid phase sequencing method[2] using streptavidin coated magnetic beads with recombinant DNA polymerase lacking 3´to 5´exonuclease activity (proof-reading) and luminescence detection using the firefly luciferase enzyme.
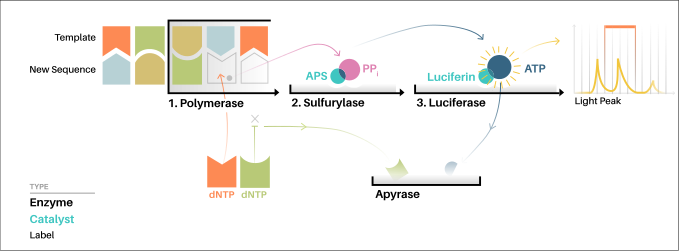
In this alternative method, an additional enzyme apyrase is introduced to remove nucleotides that are not incorporated by the DNA polymerase.
This enabled the enzyme mixture including the DNA polymerase, the luciferase and the apyrase to be added at the start and kept throughout the procedure, thus providing a simple set-up suitable for automation.
A third microfluidic variant of the pyrosequencing method was described in 2005[5] by Jonathan Rothberg and co-workers at the company 454 Life Sciences.
[6] For the solution-based version of pyrosequencing, the single-strand DNA (ssDNA) template is hybridized to a sequencing primer and incubated with the enzymes DNA polymerase, ATP sulfurylase, luciferase and apyrase, and with the substrates adenosine 5´ phosphosulfate (APS) and luciferin.
This can make the process of genome assembly more difficult, particularly for sequences containing a large amount of repetitive DNA.