Systems biology
The distinction between the two paradigms is referred to in these quotations: "the reductionist approach has successfully identified most of the components and many of the interactions but, unfortunately, offers no convincing concepts or methods to understand how system properties emerge ... the pluralism of causes and effects in biological networks is better addressed by observing, through quantitative measures, multiple components simultaneously and by rigorous data integration with mathematical models."
(Denis Noble)[7] As a series of operational protocols used for performing research, namely a cycle composed of theory, analytic or computational modelling to propose specific testable hypotheses about a biological system, experimental validation, and then using the newly acquired quantitative description of cells or cell processes to refine the computational model or theory.
[11] As a socioscientific phenomenon defined by the strategy of pursuing integration of complex data about the interactions in biological systems from diverse experimental sources using interdisciplinary tools and personnel.
In 2000, the Institute for Systems Biology was established in Seattle in an effort to lure "computational" type people who it was felt were not attracted to the academic settings of the university.
[citation needed] In 2012 the first whole-cell model of Mycoplasma genitalium was achieved by the Covert Laboratory at Stanford University.
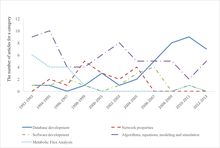
[17][18] Other early precursors that focused on the view that biology should be analyzed as a system, rather than a simple collection of parts, were Metabolic Control Analysis, developed by Henrik Kacser and Jim Burns[19] later thoroughly revised,[20] and Reinhart Heinrich and Tom Rapoport,[21] and Biochemical Systems Theory developed by Michael Savageau.
[22][23][24] According to Robert Rosen in the 1960s, holistic biology had become passé by the early 20th century, as more empirical science dominated by molecular chemistry had become popular.
[18] Echoing him forty years later in 2006 Kling writes that the success of molecular biology throughout the 20th century had suppressed holistic computational methods.
(i.e., telomere length variation); epigenomics/epigenetics, organismal and corresponding cell specific transcriptomic regulating factors not empirically coded in the genomic sequence.
); transcriptomics, organismal, tissue or whole cell gene expression measurements by DNA microarrays or serial analysis of gene expression; interferomics, organismal, tissue, or cell-level transcript correcting factors (i.e., RNA interference), proteomics, organismal, tissue, or cell level measurements of proteins and peptides via two-dimensional gel electrophoresis, mass spectrometry or multi-dimensional protein identification techniques (advanced HPLC systems coupled with mass spectrometry).
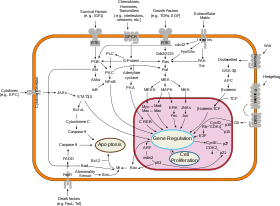
Significant efforts in computational systems biology of cancer have been made in creating realistic multi-scale in silico models of various tumours.
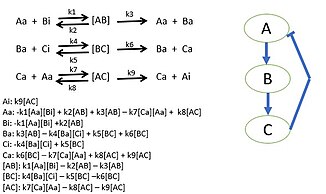
[35][36][37][38] For instance, a cellular network can be modelled mathematically using methods coming from chemical kinetics[39] and control theory.
Due to the large number of parameters, variables and constraints in cellular networks, numerical and computational techniques are often used (e.g., flux balance analysis).
Additionally, as much of the information comes from different fields, the development of syntactically and semantically sound ways of representing biological models is needed.
[45][43] The use of constraint-based reconstruction and analysis (COBRA) methods has become popular among systems biologists to simulate and predict the metabolic phenotypes, using genome-scale models.