Myc
[2] Constitutive upregulation of Myc genes have also been observed in carcinoma of the cervix, colon, breast, lung and stomach.
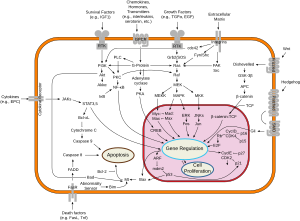
[6] In the human genome, C-myc is located on chromosome 8 and is believed to regulate expression of 15% of all genes[7] through binding on enhancer box sequences (E-boxes).
This causes c-Myc to be placed downstream of the highly active immunoglobulin (Ig) promoter region, leading to overexpression of Myc.
Myc proteins are transcription factors that activate expression of many pro-proliferative genes through binding enhancer box sequences (E-boxes) and recruiting histone acetyltransferases (HATs).
Myc-nick is a cytoplasmic form of Myc produced by a partial proteolytic cleavage of full-length c-Myc and N-Myc.
A large body of evidence shows that Myc genes and proteins are highly relevant for treating tumors.
[26][27] BET inhibitors have been used to successfully block Myc function in pre-clinical cancer models and are currently being evaluated in clinical trials.
[28] MYC expression is controlled by a wide variety of noncoding RNAs, including miRNA, lncRNA, and circRNA.
[30] MYC-R in DLBCL/HGBCL is believed to arise through the aberrant activity of activation-induced cytidine deaminase (AICDA), which facilitates somatic hypermutation (SHM) and class-switch recombination (CSR).
These breakpoints may occur within the so-called “genic cluster,” a region spanning approximately 1.5 kb upstream of the transcription start site, as well as the first exon and intron of MYC.
When MYC-R occur, two types of signals can be observed: There is a large variability in the interpretation of unbalanced MYC BAP results among the scientists, which can impact diagnostic classification and therapeutic management of the patients.
To study the mechanism of tumorigenesis in Burkitt lymphoma by mimicking expression pattern of Myc in these cancer cells, transgenic mouse models were developed.
Myc gene placed under the control of IgM heavy chain enhancer in transgenic mice gives rise to mainly lymphomas.
The study by John M. Sedivy and others used Cre-Loxp -recombinase to knockout one copy of Myc and this resulted in a "Haplo-insufficient" genotype noted as Myc+/-.
The phenotypes seen oppose the effects of normal aging and are shared with many other long-lived mouse models such as CR (calorie restriction) ames dwarf, rapamycin, metformin and resveratrol.
One study found that Myc and p53 genes were key to the survival of chronic myeloid leukaemia (CML) cells.
[42] In particular, long-term hematopoietic stem cells (LT-HSCs) express low levels of c-Myc, ensuring self-renewal.
Enforced expression of c-Myc in LT-HSCs promotes differentiation at the expense of self-renewal, resulting in stem cell exhaustion.