Nearest-neighbor chain algorithm
Both of these kinds of analysis can be seen, for instance, in the application of hierarchical clustering to biological taxonomy.
This analysis simultaneously gives a multi-scale grouping of the organisms of the present age, and aims to accurately reconstruct the branching process or evolutionary tree that in past ages produced these organisms.
[3] In agglomerative clustering methods, the input also includes a distance function defined on the points, or a numerical measure of their dissimilarity.
However, unlike the distances in a metric space, it is not required to satisfy the triangle inequality.
[2] The bottleneck of this greedy algorithm is the subproblem of finding which two clusters to merge in each step.
Known methods for repeatedly finding the closest pair of clusters in a dynamic set of clusters either require superlinear space to maintain a data structure that can find closest pairs quickly, or they take greater than linear time to find each closest pair.
In this way, it avoids the problem of repeatedly finding closest pairs.
Every cluster is only ever added once to the stack, because when it is removed again it is immediately made inactive and merged.
[2] Each of these iterations may spend time scanning as many as n − 1 inter-cluster distances to find the nearest neighbor.
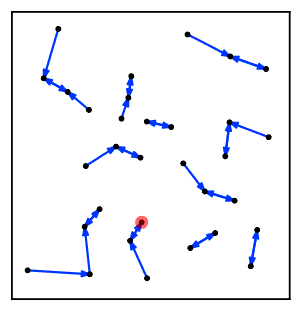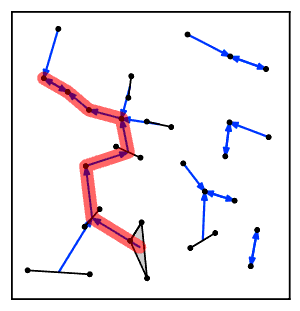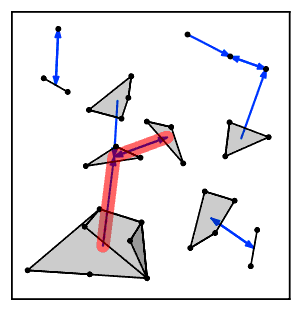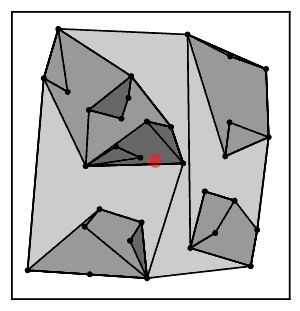
[2] For the algorithm to be correct, it must be the case that popping and merging the top two clusters from the algorithm's stack preserves the property that the remaining clusters on the stack form a chain of nearest neighbors.
Both of these properties depend on the specific choice of how to measure the distance between clusters.
[2] The correctness of this algorithm relies on a property of its distance function called reducibility.
First, it can be shown using this property that, at each step of the algorithm, the clusters on the stack S form a valid chain of nearest neighbors, because whenever a nearest neighbor becomes invalidated it is immediately removed from the stack.
of the two clusters, it has the simpler formula allowing it to be computed in constant time per distance calculation.
coefficient of one of the two other terms, leaving a positive value added to the weighted average of the other two distances.
Similarly, average-distance clustering uses the average pairwise distance as the dissimilarity.
Maintaining this array over the course of the clustering algorithm takes time and space O(n2).
[12] The same O(n2) time and space bounds can also be achieved in a different way, by a technique that overlays a quadtree-based priority queue data structure on top of the distance matrix and uses it to perform the standard greedy clustering algorithm.
[4] However, the nearest-neighbor chain algorithm matches its time and space bounds while using simpler data structures.
(Single-linkage also obeys a Lance–Williams formula,[9][11] but with a negative coefficient from which it is more difficult to prove reducibility.)
As with complete linkage and average distance, the difficulty of calculating cluster distances causes the nearest-neighbor chain algorithm to take time and space O(n2) to compute the single-linkage clustering.
However, the single-linkage clustering can be found more efficiently by an alternative algorithm that computes the minimum spanning tree of the input distances using Prim's algorithm, and then sorts the minimum spanning tree edges and uses this sorted list to guide the merger of pairs of clusters.
Within Prim's algorithm, each successive minimum spanning tree edge can be found by a sequential search through an unsorted list of the smallest edges connecting the partially constructed tree to each additional vertex.
This choice saves the time that the algorithm would otherwise spend adjusting the weights of vertices in its priority queue.
Nevertheless, Murtagh (1983) writes that the nearest-neighbor chain algorithm provides "a good heuristic" for the centroid method.
[2] A different algorithm by Day & Edelsbrunner (1984) can be used to find the greedy clustering in O(n2) time for this distance measure.
[5] The above presentation explicitly disallowed distances sensitive to merge order.
In particular, there exist order-sensitive cluster distances which satisfy reducibility, but for which the above algorithm will return a hierarchy with suboptimal costs.
Therefore, when cluster distances are defined by a recursive formula (as some of the ones discussed above are), care must be taken that they do not use the hierarchy in a way which is sensitive to merge order.
[14] The nearest-neighbor chain algorithm was developed and implemented in 1982 by Jean-Paul Benzécri[15] and J.