Biomolecular structure
Biomolecular structure is the intricate folded, three-dimensional shape that is formed by a molecule of protein, DNA, or RNA, and that is important to its function.
The structure of these molecules may be considered at any of several length scales ranging from the level of individual atoms to the relationships among entire protein subunits.
This useful distinction among scales is often expressed as a decomposition of molecular structure into four levels: primary, secondary, tertiary, and quaternary.
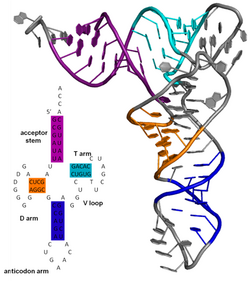
This leads to several recognizable domains of protein structure and nucleic acid structure, including such secondary-structure features as alpha helixes and beta sheets for proteins, and hairpin loops, bulges, and internal loops for nucleic acids.
The terms primary, secondary, tertiary, and quaternary structure were introduced by Kaj Ulrik Linderstrøm-Lang in his 1951 Lane Medical Lectures at Stanford University.
Famous examples include the Rho-independent terminator stem loops and the transfer RNA (tRNA) cloverleaf.
[11] However, a second mutation that reduces another morphogenetic component (e.g. in the base plate or head of the phage) could in some cases restore a balance such that a higher proportion of the virus particles produced are able to function.
[12] Protein and nucleic acid structures can be determined using either nuclear magnetic resonance spectroscopy (NMR) or X-ray crystallography or single-particle cryo electron microscopy (cryoEM).
The first published reports for DNA (by Rosalind Franklin and Raymond Gosling in 1953) of A-DNA X-ray diffraction patterns—and also B-DNA—used analyses based on Patterson function transforms that provided only a limited amount of structural information for oriented fibers of DNA isolated from calf thymus.
[14][15] An alternate analysis was then proposed by Wilkins et al. in 1953 for B-DNA X-ray diffraction and scattering patterns of hydrated, bacterial-oriented DNA fibers and trout sperm heads in terms of squares of Bessel functions.
[18] Their corresponding X-ray diffraction & scattering patterns are characteristic of molecular paracrystals with a significant degree of disorder (over 20%),[19][20] and the structure is not tractable using only the standard analysis.
In contrast, the standard analysis, involving only Fourier transforms of Bessel functions[21] and DNA molecular models, is still routinely used to analyze A-DNA and Z-DNA X-ray diffraction patterns.
A common problem for researchers working with RNA is to determine the three-dimensional structure of the molecule given only the nucleic acid sequence.
Secondary structure of small nucleic acid molecules is determined largely by strong, local interactions such as hydrogen bonds and base stacking.