Regulatory sequence
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism.
Regulation of gene expression is an essential feature of all living organisms and viruses.
In DNA, regulation of gene expression normally happens at the level of RNA biosynthesis (transcription).
[3] These cis-regulatory sequences include enhancers, silencers, insulators and tethering elements.
[4] Among this constellation of sequences, enhancers and their associated transcription factor proteins have a leading role in the regulation of gene expression.
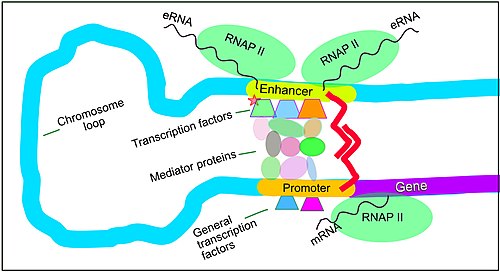
[6] In a study of brain cortical neurons, 24,937 loops were found, bringing enhancers to promoters.
[6] The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene.
Mediator (coactivator) (a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (RNAP II) enzyme bound to the promoter.
[10] Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two eRNAs as illustrated in the Figure.
[18] Transcription factors are proteins that bind to specific DNA sequences in order to regulate the expression of a given gene.
An EGR1 transcription factor binding site is frequently located in enhancer or promoter sequences.
[21] Expression of EGR1 in various types of cells can be stimulated by growth factors, neurotransmitters, hormones, stress and injury.
[26] The double-strand break introduced by TOP2B apparently frees the part of the promoter at an RNA polymerase–bound transcription start site to physically move to its associated enhancer.
[27] TOP1 causes single-strand breaks in particular enhancer DNA regulatory sequences when signaled by a specific enhancer-binding transcription factor.