DNA demethylation
[4] After surgery, demethylations are found in peripheral blood mononuclear cells at sites annotated to immune system genes.
[6] During global DNA hypomethylation of tumor genomes, there is a minor to moderate reduction of the number of methylated cytosines (5mC) amounting to a loss of about 5% to 20% on average of the 5mC bases.
[citation needed] After fertilization, the paternal chromosome is almost completely demethylated in six hours by an active process, before DNA replication (blue line in Figure).
As reviewed by Howell et al.,[8] DNMT1o is sequestered in the cytoplasm of mature oocytes and in 2-cell and 4-cell embryos, but at the 8-cell stage is only present in the nucleus.
It appears that demethylation of the maternal chromosomes largely takes place by blockage of the methylating enzyme DNMT1o from entering the nucleus except briefly at the 8 cell stage.
The morula (at the 16 cell stage), has only a small amount of DNA methylation (black line in Figure).
As reviewed by Messerschmidt et al.,[11] the majority of PGCs are arrested in the G2 phase of the cell cycle, while they migrate toward the hindgut during embryo days 7.5 to 8.5.
One particular TET enzyme, TET1, and TDG are present at high levels from embryo day 9.5 to 13.5,[12] and are employed in active demethylation during gametogenesis.
[11] PGC genomes display the lowest levels of DNA methylation of any cells in the entire life cycle of the mouse at embryonic day 13.5.
[13] Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature.
Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time.
Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory.
At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated.
There were 1,223 differentially methylated genes in the anterior cingulate cortex of mice four weeks after contextual fear conditioning.
[16] During formation of a cancer there is an average reduction of the number of methylated cytosines of about 5% to 20%,[7] or about 840,00 to 3.4 million demethylations of CpG sites.
The enzymes needed for reprogramming are recruited to genome sites that require demethylation or methylation.
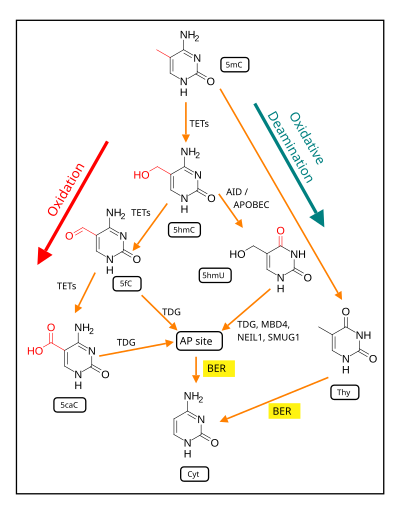
Demethylation of 5-methylcytosine to generate 5-hydroxymethylcytosine (5hmC) very often initially involves oxidation of 5mC (see Figure in this section) by ten-eleven translocation methylcytosine dioxygenases (TET enzymes).
The dominant TET1 isoform in most somatic tissues, at least in the mouse, arises from alternative promoter usage which gives rise to a short transcript and a truncated protein designated TET1s.
[24] The CXXC domain of the full-length TET3, which is the predominant form expressed in neurons, binds most strongly to CpGs where the C was converted to 5-carboxycytosine (5caC).
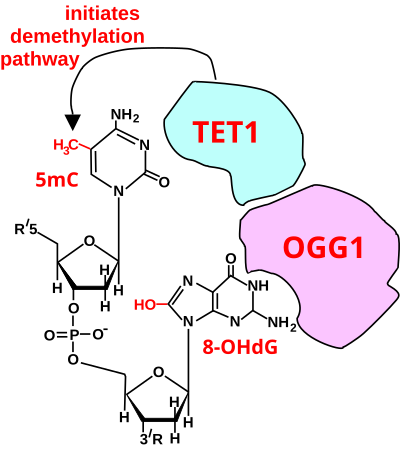
[22] For a TET enzyme to initiate demethylation it must first be recruited to a methylated CpG site in DNA.
OGG1 proteins bind to oxidatively damaged DNA with a half maximum time of about 6 seconds.
When human mammary epithelial cells (MCF-10A) were treated with H2O2, 8-OHdG increased in DNA by 3.5-fold and this caused about 80% demethylation of the 5-methylcytosines in the MCF-10A genome.
[33] The defining characteristic of IEGs is the rapid and transient up-regulation—within minutes—of their mRNA levels independent of protein synthesis.
[34] This expression is linked to control of cognition, emotional response, social behavior and sensitivity to reward.
Within the genome, 5hmC is located at transcriptionally active genes, regulatory elements and chromatin associated complexes.
In this Figure, the 8-OHdG is left in the DNA, since it may have been present when OGG1 attracted TET1 to the CpG site with a methylated cytosine.
[39] As reviewed by Fernandes et al.,[40] in rats, exercise enhances the hippocampus expression of the gene Bdnf, which has an essential role in memory formation.
[19] In a panel of healthy adults, negative associations were found between total DNA methylation and exposure to traffic related air pollution.
DNA methylation levels were associated both with recent and chronic exposure to Black Carbon as well as benzene.
DNA demethylation in mature mammalian neurons removes barriers to axonal regeneration.