Ribonucleotide reductase
[1][2] It catalyzes this formation by removing the 2'-hydroxyl group of the ribose ring of nucleoside diphosphates (or triphosphates depending on the class of RNR).
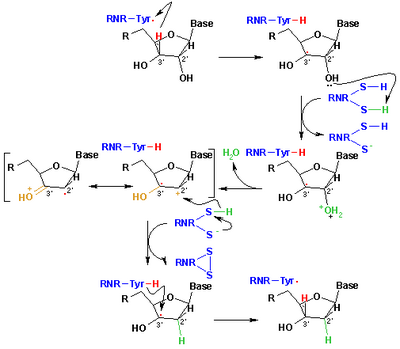
[5] A somewhat unusual feature of the RNR enzyme is that it catalyzes a reaction that proceeds via a free radical mechanism of action.
Other residues of RNR2, such as aspartate (D273), tryptophan (W48), and tyrosine (Y356) further stabilize the active-site tyrosyl radical thus allowing electron transfer.
[16] Site-directed mutations of the RNR primary structure indicate that all residues cited above participate in the long distance transfer of the free radical to the active site.
Following a single reduction, RNR requires electrons donated from the dithiol groups of the protein thioredoxin.
Three classes of RNR have similar mechanisms for the reduction of NDPs, but differ in the domain that generates the free radical, the specific metal in the metalloprotein structure, and the electron donors.
[10] Class I reductases use an iron center with ferrous to ferric conversion to generate a tyrosyl free radical.
Class III reductases use a glycine radical generated with the help of an S-adenosyl methionine and an iron sulphur center.
At the end of this step, a radical anionic disulfide bridge and the closed-shell ketone intermediate 4 are obtained.
[10] In addition to controlling activity, the allosteric mechanism also regulates the substrate specificity and ensures the enzyme produces an equal amount of each dNTP for DNA synthesis.
Class IB reductases are not inhibited by dATP because they lack approximately 50 N-terminal amino acids required for the allosteric activity site.
[23] Additionally, it is important that the activity of ribonucleotide reductase be under transcriptional and post-transcriptional control because the synthesis of damage-free DNA relies on a balanced pool of deoxyribonucleotides.
[24] Eukaryotic cells with class IA reductases have a mechanism of negative control to turn off synthesis of dNTPs as they accumulate.
Changes within this p53 induced R2 homolog can cause depletion in mitochondrial DNA and consequently p53R2 serves a major factor in dNTP supply.
However, they lack enzymatic activity due probably to the elimination of residues involved in the transfer of the free radical electron from the RNR2 to the RNR1 subunit.
[30] Small peptides can specifically inhibit the RNR2 subunits from binding with RNR1 when they share a significant similarity with the normal RNR2 C-terminus.
When a 7 amino acid oligomer (GAVVNDL) truncated from the C-terminus of the RNR2 subunit was used in competition assays, it prevented the normal RNR2 from forming an enzymatically active complex with RNR1.
[36] A combination therapy approach (BILD 1633 and acyclovir) is more effective to heal topical lesions in mice.
These data suggest that small peptide inhibitors that compete with RNR2 for binding to RNR1 are useful in preventing the spread of HSV.