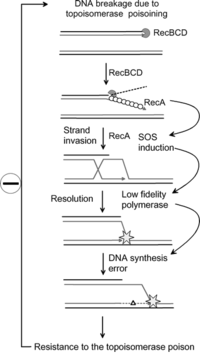
SOS response
It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.
[3][4] Later, by characterizing the phenotypes of mutagenised E. coli, she and post doctoral student Miroslav Radman detailed the SOS response to UV radiation in bacteria.
Under normal conditions, LexA binds to a 20-bp consensus sequence (the SOS box) in the operator region for those genes.
The SOS dependent tisB-istR toxin-antitoxin system has, for example, been linked to DNA damage-dependent persister cell induction.
A lactose analog is added to the bacteria, which is then degraded by beta-galactosidase, thereby producing a colored compound which can be measured quantitatively through spectrophotometry.
The degree of color development is an indirect measure of the beta-galactosidase produced, which itself is directly related to the amount of DNA damage.
[13] Commercial kits which measures the primary response of the E. coli cell to genetic damage are available and may be highly correlated with the Ames Test for certain materials.