Nucleotide excision repair
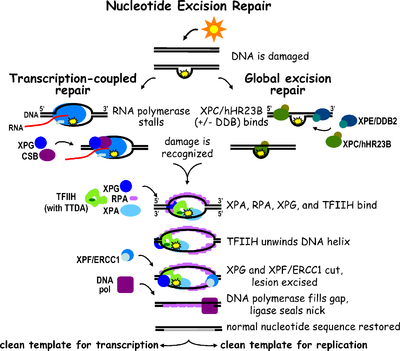
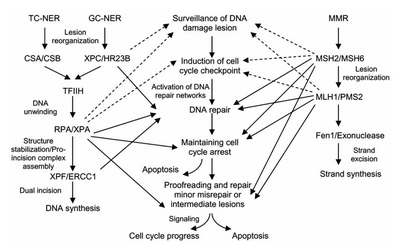
The two subpathways differ in how they recognize DNA damage but they share the same process for lesion incision, repair, and ligation.
XPA, XPB, XPC, XPD, XPE, XPF, and XPG all derive from хeroderma pigmentosum and CSA and CSB represent proteins linked to Cockayne syndrome.
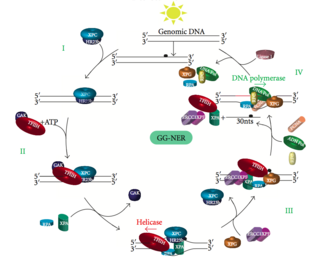
This pathway employs several "damage sensing" proteins including the DNA-damage binding (DDB) and XPC-Rad23B complexes that constantly scan the genome and recognize helix distortions: the XPC-Rad23B complex is responsible for distortion recognition, while DDB1 and DDB2 (XPE) can also recognize some types of damage caused by UV light.
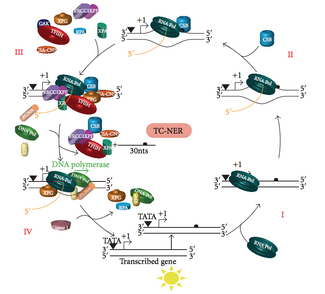
Mutations in TC-NER machinery are responsible for multiple genetic disorders including: Transcription factor II H (TFIIH) is the key enzyme involved in dual excision.
The small, excised, damage-containing DNA (sedDNA) oligonucleotides are initially released from the duplex in complex with TFIIH but then dissociate in an ATP-dependent manner and become bound to replication protein A (RPA).
Inhibition of gap filling DNA synthesis and ligation results in an accumulation of RPA-bound sedDNAs in the cell.
This allows DNA polymerases implicated in repair (δ, ε and/or κ) to copy the undamaged strand via translocation.
First, a UvrA-UvrB complex scans the DNA, with the UvrA subunit recognizing distortions in the helix, caused for example by pyrimidine dimers.
When the complex recognizes such a distortion, the UvrA subunit leaves and an UvrC protein comes in and binds to the UvrB monomer and, hence, forms a new UvrBC dimer.
DNA helicase II (sometimes called UvrD) then comes in and removes the excised segment by actively breaking the hydrogen bonds between the complementary bases.
TRCF also recruits the Uvr(A)BC nucleotide excision repair machinery by direct physical interaction with the UvrA subunit.
[7] If located in NER genes or regulatory sequences, such mutations can negatively affect DNA repair capacity resulting in an increase likelihood of cancer development.
Research has shown that a biallelic poly (AT) insertion/deletion polymorphism in Intron 9 of XPC is associated with increased risk for skin, breast and prostate cancers,[12][13][14] especially in North Indian populations.
The study of a hereditary cancer, xeroderma pigmentosum has helped identify several genes which encode proteins in the NER pathway, two of which are XPC and XPD.
XP is caused by a homozygous deficiency in UV DNA damage repair (GG-NER) which increases the patients' risk of skin cancer by 1000-fold.
In heterozygous patients, the risk of cancer is sporadic but can be predicted based on analytical assessment of polymorphisms in XP related DNA repair genes purified from lymphocytes.
[15][16][17] In humans and mice, germline mutation in genes employed in NER cause features of premature aging.
These features may include sensorineural deafness, retinal degeneration, white matter hypomethylation, central nervous system calcification, reduced stature, and cachexia (loss of subcutaneous fat tissue).
[24] An ERCC5(XPG) mutant mouse model presents features of premature aging including cachexia and osteoporosis with pronounced degenerative phenotypes in both liver and brain.