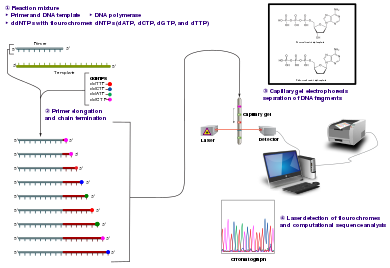
Sanger sequencing
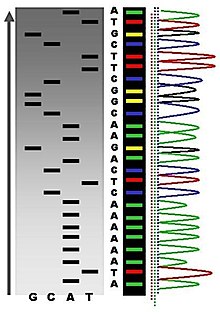
In the image on the right, X-ray film was exposed to the gel, and the dark bands correspond to DNA fragments of different lengths.
A dark band in a lane indicates a DNA fragment that is the result of chain termination after incorporation of a dideoxynucleotide (ddATP, ddGTP, ddCTP, or ddTTP).
Technical variations of chain-termination sequencing include tagging with nucleotides containing radioactive phosphorus for radiolabelling, or using a primer labeled at the 5' end with a fluorescent dye.
In dye-terminator sequencing, each of the four dideoxynucleotide chain terminators is labelled with fluorescent dyes, each of which emits light at different wavelengths.
Its limitations include dye effects due to differences in the incorporation of the dye-labelled chain terminators into the DNA fragment, resulting in unequal peak heights and shapes in the electronic DNA sequence trace electropherogram (a type of chromatogram) after capillary electrophoresis (see figure to the left).
[9] The accuracy of such algorithms is inferior to visual examination by a human operator, but is adequate for automated processing of large sequence data sets.
The field of public health plays many roles to support patient diagnostics as well as environmental surveillance of potential toxic substances and circulating biological pathogens.
[10] Laboratories were tasked with the rapid implementation of sequencing methods and asked to provide accurate data to assist in the decision-making models for the development of policies to mitigate spread of the virus.
Sanger sequencing is also the "gold standard" for norovirus surveillance methods for the Center for Disease Control and Prevention's (CDC) CaliciNet network.
The goal of the network is to collect sequencing data of circulating noroviruses in the United States and activate downstream action to determine the source of infection to mitigate the spread of the virus.
Base calling software such as Phred typically provides an estimate of quality to aid in trimming of low-quality regions of sequences.
[14][15] Current methods can directly sequence only relatively short (300–1000 nucleotides long) DNA fragments in a single reaction.
Adopting the Sanger method, each DNA fragment is irreversibly terminated with the incorporation of a fluorescently labeled dideoxy chain-terminating nucleotide, thereby producing a DNA “ladder” of fragments that each differ in length by one base and bear a base-specific fluorescent label at the terminal base.
Resolving DNA fragments according to differences in size and/or conformation is the most critical step in studying these features of the genome.
[16] The sequencing chip has a four-layer construction, consisting of three 100-mm-diameter glass wafers (on which device elements are microfabricated) and a polydimethylsiloxane (PDMS) membrane.
The thermal cycling (TC) unit is a 250-nanoliter reaction chamber with integrated resistive temperature detector, microvalves, and a surface heater.
The Apollo 100 platform (Microchip Biotechnologies Inc., Dublin, California)[18] integrates the first two Sanger sequencing steps (thermal cycling and purification) in a fully automated system.
The ultimate goal of high-throughput sequencing is to develop systems that are low-cost, and extremely efficient at obtaining extended (longer) read lengths.