Sphingomyelin phosphodiesterase
[3] Subsequent study found that this enzyme was the product of a distinct gene, had an optimum pH of 7.4, was dependent on Mg2+ ions for activity, and was particularly enriched in brain.
[4] However, a more recent study in bovine brain suggested the existence of multiple N-SMase isoforms with different biochemical and chromatographical properties.
However, there appears to be a high degree of evolutionary conservation from lower to higher organisms, suggesting that it may comprise a unique and distinct N-SMase.
[13] The solving of the crystal structure of the neutral sphingomyelinase from Listeria ivanovii and Bacillus cereus has allowed a fuller understanding of their enzymatic site.
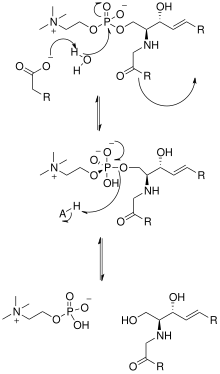
[1] The solving of the crystal structure of the neutral sphingomyelinase from Listeria ivanovii and Bacillus cereus has also shed light on their catalytic mechanisms.
The active site of SMase contains Glu and His residues that are each bound to one or two divalent metal cations, usually Co2+, Mg2+, or Ca2+ for optimum performance.
This water molecule then acts as a nucleophile and attacks the phosphate group of SM, creating a pentavalent phosphorus atom whose negative charge is stabilized by the divalent metal cations.
[1] In 2016 a model based on crystal structure of mammalian acid sphingomyelinase study was proposed whereby ASMase exists in equilibrium between open and closed forms of the saposin domain.