Type III secretion system
These T3SSs are similar as a result of convergent evolution and phylogenetic analysis supports a model in which gram-negative bacteria can transfer the T3SS gene cassette horizontally to other species.
Some of the most researched T3SSs are from species of:[citation needed] The T3SS is composed of approximately 30 different proteins, making it one of the most complex secretion systems.
It has been suggested that some non-invasive strains of gram-negative bacteria have lost the T3SS because the energetically costly system is no longer of use.
Understanding the way the T3SS works and developing drugs targeting it specifically have become an important goal of many research groups around the world since the late 1990s.
Once the whole complex is completed the system switches to secreting proteins that are intended to be delivered into host cells.
It needs to have a minimal length so that other extracellular bacterial structures (adhesins and the lipopolysaccharide layer, for instance) do not interfere with secretion.
It is important to note that many pathogenicity islands and plasmids contain elements that allow for frequent horizontal gene transfer of the island/plasmid to a new species.
Induction of secretion by external cues other than contact with host cells also takes place in vivo, in infected organisms.
Molecules present in the cecum, such as propionate and butyrate, provide a negative cue to the bacteria and inhibit secretion.
A feedback mechanism has been suggested: when the bacterium does not secrete, its effector proteins are bound to chaperones and float in the cytoplasm.
It has been previously suggested that the needle itself is capable of puncturing a hole in the host cell membrane; this theory has been refuted.
Some translocators serve a double role; after they participate in pore formation they enter the cell and act as bona fide effectors.
In order for this to happen the bacterial effectors manipulate the actin polymerization machinery of the host cell.
[16] It was later shown that IpaB achieves this by interacting with caspase 1, a major regulatory protein in eukaryotic cells.
[18] TAL effector-DNA recognition has recently been demonstrated to comprise a simple code[19][20] and this has greatly improved the understanding of how these proteins can alter the transcription of genes in the host plant cells.
However, numerous issues regarding the system remain unresolved: Since the beginning of the 1990s new T3SS proteins are being found in different bacterial species at a steady rate.
For example, the proteins SicA, IpgC and SycD are homologs from Salmonella, Shigella and Yersinia, respectively, but the last letter (the "serial number") in their name does not show that.
The following table shows some of these key proteins in four T3SS-containing bacteria: The isolation of large, fragile, hydrophobic membrane structures from cells has constituted a challenge for many years.
[29] For the isolation, bacteria are grown in a large volume of liquid growth medium until they reach log phase.
They are then centrifuged; the supernatant (the medium) is discarded and the pellet (the bacteria) is resuspended in a lysis buffer typically containing lysozyme and sometimes a detergent such as LDAO or Triton X-100.
Type III effectors were known since the beginning of the 1990s, but the way in which they are delivered into host cells was a complete mystery.
The homology between many flagellar and T3SS proteins led researchers to suspects the existence of an outer T3SS structure similar to flagella.
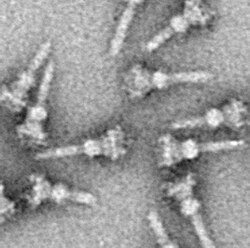
The identification and subsequent isolation of the needle structure enabled researchers to: As with almost all proteins, the visualization of T3SS NCs is only possible with electron microscopy.
The model also revealed an extended amino-terminal domain that is positioned on the surface of the needle, while the highly conserved carboxy terminus points towards the lumen.
The bands that appear after staining can be individually excised from the gel and analyzed using protein sequencing and mass spectrometry.
Alternatively, isolated NCs can be directly analyzed by mass spectrometry, without prior electrophoresis, in order to obtain a complete picture of the NC proteome.
Examples of possible influences: A few compounds have been discovered that inhibit the T3SS in gram-negative bacteria, including the guadinomines which are naturally produced by Streptomyces species.
[39] Aurodox, an antibiotic capable of inhibiting the translation of T3SS proteins has been shown to able to prevent T3SS effectors in vitro and in animal models[40][41]