Proteome
A proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time.
It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions.
A cellular proteome is the collection of proteins found in a particular cell type under a particular set of environmental conditions such as exposure to hormone stimulation.
Proteomic studies have been used in order to identify the likelihood of metastasis in bladder cancer cell lines KK47 and YTS1 and were found to have 36 unregulated and 74 down regulated proteins.
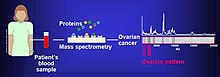
[8] The analysis of ovarian cancer cell lines showed that putative biomarkers for ovarian cancer include "α-enolase (ENOA), elongation factor Tu, mitochondrial (EFTU), glyceraldehyde-3-phosphate dehydrogenase (G3P), stress-70 protein, mitochondrial (GRP75), apolipoprotein A-1 (APOA1), peroxiredoxin (PRDX2) and annexin A (ANXA)".
Proteins that showed a differential expression were involved in processes such as transcription, apoptosis and cell proliferation/differentiation among others.
Marc Wilkins coined the term proteome [13] in 1994 in a symposium on "2D Electrophoresis: from protein maps to genomes" held in Siena in Italy.
Wilkins used the term to describe the entire complement of proteins expressed by a genome, cell, tissue or organism.
[18] “Proteomic constraint” proposes that modulators of mutation rates such as DNA repair genes are subject to selection pressure proportional to the amount of information in a genome.
For 546,000 Swiss-Prot proteins, 44–54% of the proteome in eukaryotes and viruses was found to be "dark", compared with only ~14% in archaea and bacteria.
The Human Proteome Map currently (October 2020) claims 17,294 proteins and ProteomicsDB 15,479, using different criteria.
On October 16, 2020, the HPP published a high-stringency blueprint [21] covering more than 90% of the predicted protein coding genes.
Tandem mass spectrometry, on the other hand, can get sequence information from individual peptides by isolating them, colliding them with a non-reactive gas, and then cataloguing the fragment ions produced.
This study profiled 30 histologically normal human samples resulting in the identification of proteins coded by 17,294 genes.
In 2022, a large-scale collaboration between EMBL-EBI and DeepMind provided predicted structures for over 200 million proteins from across the tree of life.
The organization ELIXIR has selected the protein atlas as a core resource due to its fundamental importance for a wider life science community.
This is an analytical limit that may possibly be a barrier for the detections of proteins with ultra low concentrations.