Anti-thrombin aptamers
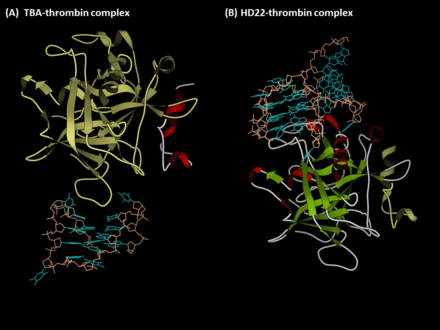
The first anti-thrombin aptamer, TBA, was generated through via SELEX (Systematic Evolution of Ligands by Exponential Enrichment) technology in 1992 by L.C.
[1] A second thrombin-binding aptamer, HD22, recognizes thrombin exosite II and was discovered in 1997 by NeXstar (now Gilead Sciences).
[2] These two aptamers have high affinity and good specificity and have been widely studied and used for the development of aptamer-based therapeutics and diagnostics.
The dissociation constant of TBA-thrombin has been reported in nano-molar range, and TBA does not interact with other plasma proteins or thrombin analogues (e.g., gamma-thrombin).
[3] As a result, TBA has been used as a short-term anti-coagulant designed for the application in the coronary artery bypass graft surgery, and its optimized form (NU172) is now under the phase II of clinical trial by ARCA Biopharma (NCT00808964).
[4] Also, due to its high affinity and specificity, a variety of sensors was coupled with TBA and developed for thrombosis diagnostics.
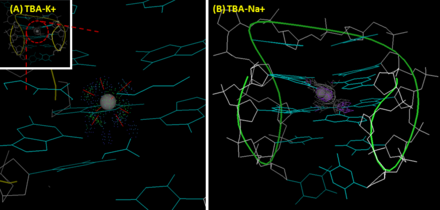
[8] The melting temperature of TBA's G-quadruplex (measuring the intensity change of the peak at 295 nm by CD) in the presence of sodium ion and potassium are 24 °C and 53 °C, respectively.
In contrast, due to its small size, sodium ion can only interacts with four rather than eight oxygen atoms of two G-tetrad planes, and accordingly has two alternative position in the cavity.
In the ion-deficient condition, thrombin helps TBA form into a stable G-quadruplex structure from a randomized coil, which results in conformational change.
For this purpose, TBA is usually mounted with an additional sequence with a FRET (Förster resonance energy transfer) pair to form a transient duplex structure.
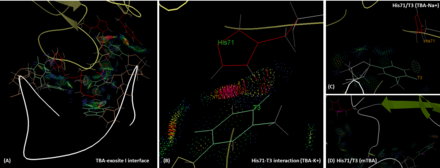
[10] TBA is bound to the exosite I of thrombin majorly via its two TT loops (T3, T4 and T12, T13) through polar and hydrophobic interactions.
However, in the presence of sodium ion, the hydrogen bonding between T3 and His71 is lost, and the intermolecular distance is longer than that in the potassium case.
[16] TBA entered the phase I clinical trial for coronary artery bypass graft surgery by Archemix and Nuvelo (now ARCA Biopharma) around 2005.
[17] Thus, the companies redesigned the sequence of TBA and developed a second-generation 26-mer DNA aptamer known as NU172, which is now under phase II clinical trial.
Since the exosite II is a positively charged motif, it creates many ion pairs with the HD22 backbone especially in the duplex region.
Moreover, Interacting with thrombin improves the thermal stability of HD22 structure, and results in the increase of melting temperature (from 36 to 48 °C).