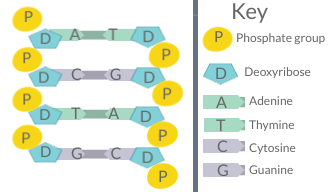
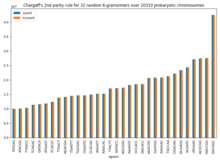
Chargaff's rules
The first rule holds that a double-stranded DNA molecule, globally has percentage base pair equality: A% = T% and G% = C%.
The rigorous validation of the rule constitutes the basis of Watson–Crick base pairs in the DNA double helix model.
The origin of the deviation from Chargaff's rule in the organelles has been suggested to be a consequence of the mechanism of replication.
For reasons that are not yet clear the strands tend to exist longer in single form in mitochondria than in chromosomal DNA.
[citation needed][dubious – discuss] Chargaff's second rule appears to be the consequence of a more complex parity rule: within a single strand of DNA any oligonucleotide (k-mer or n-gram; length ≤ 10) is present in equal numbers to its reverse complementary nucleotide.
[11] Albrecht-Buehler has suggested that this rule is the consequence of genomes evolving by a process of inversion and transposition.
Chargaff's second parity rule appears to be extended from the nucleotide-level to populations of codon triplets, in the case of whole single-stranded Human genome DNA.
[12] A kind of "codon-level second Chargaff's parity rule" is proposed as follows: Examples — computing whole human genome using the first codons reading frame provides: In 2020, it is suggested that the physical properties of the dsDNA (double stranded DNA) and the tendency to maximum entropy of all the physical systems are the cause of Chargaff's second parity rule.