Genetic code
Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time.
[2] Soviet-American physicist George Gamow was the first to give a workable scheme for protein synthesis from DNA.
[5] In 1954, Gamow created an informal scientific organisation the RNA Tie Club, as suggested by Watson, for scientists of different persuasions who were interested in how proteins were synthesised from genes.
Crick presented a type-written paper titled "On Degenerate Templates and the Adaptor Hypothesis: A Note for the RNA Tie Club"[9] to the members of the club in January 1955, which "totally changed the way we thought about protein synthesis", as Watson recalled.
[11] The Crick, Brenner, Barnett and Watts-Tobin experiment first demonstrated that codons consist of three DNA bases.
[12] They used a cell-free system to translate a poly-uracil RNA sequence (i.e., UUUUU...) and discovered that the polypeptide that they had synthesized consisted of only the amino acid phenylalanine.
[16] Extending this work, Nirenberg and Philip Leder revealed the code's triplet nature and deciphered its codons.
In these experiments, various combinations of mRNA were passed through a filter that contained ribosomes, the components of cells that translate RNA into protein.
Even models are proposed that predict "entry points" for synthetic amino acid invasion of the genetic code.
[24] In 2015 N. Budisa, D. Söll and co-workers reported the full substitution of all 20,899 tryptophan residues (UGG codons) with unnatural thienopyrrole-alanine in the genetic code of the bacterium Escherichia coli.
[26][27] In 2017, researchers in South Korea reported that they had engineered a mouse with an extended genetic code that can produce proteins with unnatural amino acids.
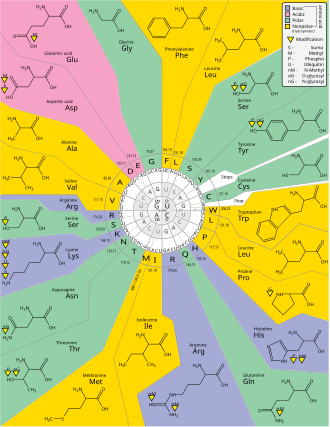
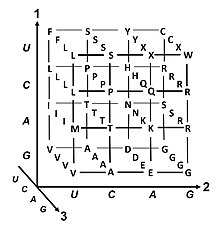
The most common start codon is AUG, which is read as methionine or as formylmethionine (in bacteria, mitochondria, and plastids).
[33] The three stop codons have names: UAG is amber, UGA is opal (sometimes also called umber), and UAA is ochre.
These errors, mutations, can affect an organism's phenotype, especially if they occur within the protein coding sequence of a gene.
These mutations usually result in a completely different translation from the original, and likely cause a stop codon to be read, which truncates the protein.
[47] In large populations of asexually reproducing organisms, for example, E. coli, multiple beneficial mutations may co-occur.
For example, the amino acid leucine is specified by YUR or CUN (UUA, UUG, CUU, CUC, CUA, or CUG) codons (difference in the first or third position indicated using IUPAC notation), while the amino acid serine is specified by UCN or AGY (UCA, UCG, UCC, UCU, AGU, or AGC) codons (difference in the first, second, or third position).
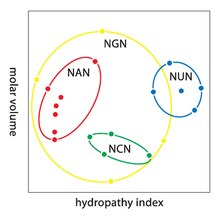
[49] A practical consequence of redundancy is that errors in the third position of the triplet codon cause only a silent mutation or an error that would not affect the protein because the hydrophilicity or hydrophobicity is maintained by equivalent substitution of amino acids; for example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids.
The genetic code is so well-structured for hydropathicity that a mathematical analysis (Singular Value Decomposition) of 12 variables (4 nucleotides x 3 positions) yields a remarkable correlation (C = 0.95) for predicting the hydropathicity of the encoded amino acid directly from the triplet nucleotide sequence, without translation.
[53] In some proteins, non-standard amino acids are substituted for standard stop codons, depending on associated signal sequences in the messenger RNA.
[60] Surprisingly, variations in the interpretation of the genetic code exist also in human nuclear-encoded genes: In 2016, researchers studying the translation of malate dehydrogenase found that in about 4% of the mRNAs encoding this enzyme the stop codon is naturally used to encode the amino acids tryptophan and arginine.
[66] This type of recoding is induced by a high-readthrough stop codon context[67] and it is referred to as functional translational readthrough.
[69] The most extreme variations occur in certain ciliates where the meaning of stop codons depends on their position within mRNA.
When close to the 3' end they act as terminators while in internal positions they either code for amino acids as in Condylostoma magnum[70] or trigger ribosomal frameshifting as in Euplotes.
For example, the program FACIL infers a genetic code by searching which amino acids in homologous protein domains are most often aligned to every codon.
[57] As of January 2022, the most complete survey of genetic codes is done by Shulgina and Eddy, who screened 250,000 prokaryotic genomes using their Codetta tool.
Despite the NCBI already providing 27 translation tables, the authors were able to find new 5 genetic code variations (corroborated by tRNA mutations) and correct several misattributions.
[80] A hypothetical randomly evolved genetic code further motivates a biochemical or evolutionary model for its origin.