Chromatin remodeling
[1] Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency.
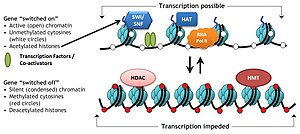
Such condensed structure occludes many DNA regulatory regions, not allowing them to interact with transcriptional machinery proteins and regulate gene expression.
These enzymatic modifications include acetylation, methylation, phosphorylation, and ubiquitination and primarily occur at N-terminal histone tails.
On the contrary, histone acetylation relaxes chromatin condensation and exposes DNA for TF binding, leading to increased gene expression.
Cumulative evidence suggests that such code is written by specific enzymes which can (for example) methylate or acetylate DNA ('writers'), removed by other enzymes having demethylase or deacetylase activity ('erasers'), and finally readily identified by proteins ('readers') that are recruited to such histone modifications and bind via specific domains, e.g., bromodomain, chromodomain.
Although all of remodelers share common ATPase domain, their functions are specific based on several biological processes (DNA repair, apoptosis, etc.).
Chromatin remodeling plays a central role in the regulation of gene expression by providing the transcription machinery with dynamic access to an otherwise tightly packaged genome.
Further, nucleosome movement by chromatin remodelers is essential to several important biological processes, including chromosome assembly and segregation, DNA replication and repair, embryonic development and pluripotency, and cell-cycle progression.
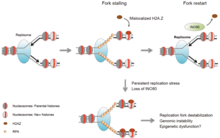
[18] γH2AX (phosphorylated on serine 139 of H2AX) was detected at 20 seconds after irradiation of cells (with DNA double-strand break formation), and half maximum accumulation of γH2AX occurred in one minute.
[18] γH2AX does not, by itself, cause chromatin decondensation, but within seconds of irradiation the protein "Mediator of the DNA damage checkpoint 1" (MDC1) specifically attaches to γH2AX.
[21] RNF8 mediates extensive chromatin decondensation, through its subsequent interaction with CHD4 protein,[22] a component of the nucleosome remodeling and deacetylase complex NuRD.
[16] Chromatin remodeling provides fine-tuning at crucial cell growth and division steps, like cell-cycle progression, DNA repair and chromosome segregation, and therefore exerts tumor-suppressor function.
Recent data indicate that chromatin inactivation mediated by HDAC and DNA methylation is a critical component of ERα silencing in human breast cancer cells.
[47][48] Senescence can arise due to age associated degradation, telomere attrition, progerias, pre-malignancies, and other forms of damage or disease.
[51] During senescence, portions of chromosomes can be exported from the nucleus for lysosomal degradation which results in greater organizational disarray and disruption of chromatin interactions.
[57][52][50][55] Specific chromatin regions, especially those around the promoters or enhancers of proliferative loci, may exhibit elevated methylation states with an overall imbalance of repressive and activating histone modifications.
[52] Additionally, upregulating histone deacetylases, such as members of the sirtuin family, can delay senescence by removing acetyl groups that contribute to greater chromatin accessibility.
[58] General loss of methylation, combined with the addition of acetyl groups results in a more accessible chromatin conformation with a propensity towards disorganization when compared to mitotically active cells.