Base excision repair
BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication.
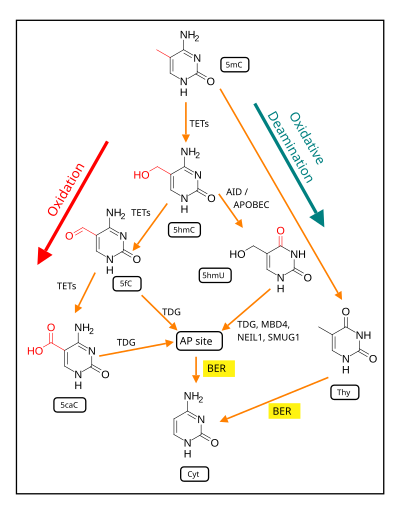
BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites.
[1] Single bases in DNA can be chemically damaged by a variety of mechanisms, the most common ones being deamination, oxidation, and alkylation.
These modifications can affect the ability of the base to hydrogen-bond, resulting in incorrect base-pairing, and, as a consequence, mutations in the DNA.
[3] Some lesions, such as oxidized or reduced AP sites, are resistant to pol β lyase activity and, therefore, must be processed by long-patch BER.
While human cells utilize both short- and long-patch BER, the yeast Saccharomyces cerevisiae was long thought to lack a short-patch pathway because it does not have homologs of several mammalian short-patch proteins, including pol β, DNA ligase III, XRCC1, and the kinase domain of PNKP.
[6] Many eukaryotes have members of both families, including the yeast Saccharomyces cerevisiae, in which Apn1 is the EndoIV homolog and Apn2 is related to ExoIII.
Besides opening AP sites, they possess 3' phosphodiesterase activity and can remove a variety of 3' lesions including phosphates, phosphoglycolates, and aldehydes.
In addition to polymerase activity, these enzymes have a lyase domain that removes the 5' dRP left behind by AP endonuclease cleavage.
In addition to its role in long-patch BER, FEN1 cleaves flaps with a similar structure during Okazaki fragment processing, an important step in lagging strand DNA replication.
[13] Defects in a variety of DNA repair pathways lead to cancer predisposition, and BER appears to follow this pattern.
[citation needed] Some examples of epimutations in base excision repair genes that occur in cancers are summarized below.
[17] This is an important repair function since about 1/3 of all intragenic single base pair mutations in human cancers occur in CpG dinucleotides and are the result of G:C to A:T transitions.
For example, nearly 50% of somatic mutations of the tumor suppressor gene p53 in colorectal cancer are G:C to A:T transitions within CpG sites.
[21] NEIL1 recognizes (targets) and removes certain oxidatively-damaged bases and then incises the abasic site via β,δ elimination, leaving 3′ and 5′ phosphate ends.
The authors suggested that low NEIL1 activity arising from reduced expression and/or mutation in NEIL1 was often involved in gastric carcinogenesis.
[25] When 8 DNA repair genes were evaluated in non-small cell lung cancer (NSCLC) tumors, 42% were hypermethylated in the NEIL1 promoter region.
[29] With contextual fear conditioning, after 24 hours, DNA isolated from the rat brain hippocampus region had 2097 differentially methylated genes, with a proportion being demethylated.
[30] As reviewed by Fernandes et al.,[31] in rats, exercise enhances the hippocampus expression of the gene Bdnf, which has an essential role in memory formation.