Genetic recombination
The information transfer may occur without physical exchange (a section of genetic material is copied from one chromosome to another, without the donating chromosome being changed) (see SDSA – Synthesis Dependent Strand Annealing pathway in Figure); or by the breaking and rejoining of DNA strands, which forms new molecules of DNA (see DHJ pathway in Figure).
RecA, the chief recombinase found in Escherichia coli, is responsible for the repair of DNA double strand breaks (DSBs).
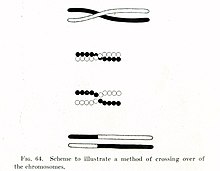
The crossover process leads to offspring having different combinations of genes from those of their parents, and can occasionally produce new chimeric alleles.
[citation needed] While in this formation, homologous sites on two chromatids can closely pair with one another, and may exchange genetic information.
[citation needed] Therefore, for genes sufficiently distant on the same chromosome, the amount of crossover is high enough to destroy the correlation between alleles.
Gene conversion has often been studied in fungal crosses[9] where the 4 products of individual meioses can be conveniently observed.
Examples include Restriction enzyme mediated integration, Gibson assembly and Golden Gate Cloning.
[10][11] These findings suggest that DNA damages arising from natural processes, such as exposure to reactive oxygen species that are byproducts of normal metabolism, are also repaired by HRR.
In bacteria, transformation is a process of gene transfer that ordinarily occurs between individual cells of the same bacterial species.
A molecular model for the mechanism of meiotic recombination presented by Anderson and Sekelsky[15] is outlined in the first figure in this article.
Recombination, in this model, is initiated by a double-strand break (or gap) shown in the DNA molecule (chromatid) at the top of the figure.
The CO type of recombination involves the intermediate formation of two "Holliday junctions" indicated in the lower right of the figure by two X-shaped structures in each of which there is an exchange of single strands between the two participating chromatids.
The NCO recombinants (illustrated on the left in the figure) are produced by a process referred to as "synthesis dependent strand annealing" (SDSA).
[16] The NCO/SDSA pathway contributes little to genetic variation, since the arms of the chromosomes flanking the recombination event remain in the parental configuration.
Thus, explanations for the adaptive function of meiosis that focus exclusively on crossing-over are inadequate to explain the majority of recombination events.
[18] Numerous RNA viruses are capable of genetic recombination when at least two viral genomes are present in the same host cell.
[21] RNA recombination appears to be a major driving force in determining genome architecture and the course of viral evolution among picornaviridae ((+)ssRNA) (e.g.
[22] In the retroviridae ((+)ssRNA)(e.g. HIV), damage in the RNA genome appears to be avoided during reverse transcription by strand switching, a form of recombination.
[25] Especially in coronaviruses, recombination may also occur even among distantly related evolutionary groups (subgenera), due to their characteristic transcription mechanism, that involves subgenomic mRNAs that are formed by template switching.
Recombination appears to occur by a copy choice mechanism in which the RdRp switches (+)ssRNA templates during negative strand synthesis.
[31] During the first months of the COVID-19 pandemic, such a recombination event was suggested to have been a critical step in the evolution of SARS-CoV-2's ability to infect humans.
The findings indicate that the 11083G > T mutation of SARS-CoV-2 spread during Diamond Princess shipboard quarantine and arose through de novo RNA recombination under positive selection pressure.
[33] SARS-CoV-2's entire receptor binding motif appeared, based on preliminary observations, to have been introduced through recombination from coronaviruses of pangolins.
Smail et al.[39] proposed that in the primordial Earth, recombination played a key role in the expansion of the initially short informational polymers (presumed to be RNA) that were the precursors to life.