Computational biology
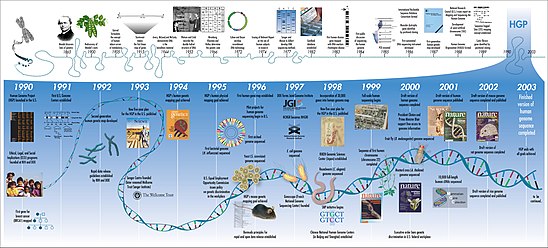
The amount of data grew exponentially by the end of the 1980s, requiring new computational methods for quickly interpreting relevant information.
[8] Today, the International Society for Computational Biology recognizes 21 different 'Communities of Special Interest', each representing a slice of the larger field.
[3] In 2000, despite a lack of initial expertise in programming and data management, Colombia began applying computational biology from an industrial perspective, focusing on plant diseases.
This research has contributed to understanding how to counteract diseases in crops like potatoes and studying the genetic diversity of coffee plants.
[10] By 2007, concerns about alternative energy sources and global climate change prompted biologists to collaborate with systems and computer engineers.
In 2009, in partnership with the University of Los Angeles, Colombia also created a Virtual Learning Environment (VLE) to improve the integration of computational biology and bioinformatics.
[11] The application of statistical models in Poland has advanced techniques for studying proteins and RNA, contributing to global scientific progress.
Polish scientists have also been instrumental in evaluating protein prediction methods, significantly enhancing the field of computational biology.
Over time, they have expanded their research to cover topics such as protein-coding analysis and hybrid structures, further solidifying Poland's influence on the development of bioinformatics worldwide.
The original formulation of computational anatomy is as a generative model of shape and form from exemplars acted upon via transformations.
It relates with shape statistics and morphometrics, with the distinction that diffeomorphisms are used to map coordinate systems, whose study is known as diffeomorphometry.
[14] These mathematical approaches have enabled the creation of databases and other methods for storing, retrieving, and analyzing biological data, a field known as bioinformatics.
[15] While current techniques focus on small biological systems, researchers are working on approaches that will allow for larger networks to be analyzed and modeled.
Sequence alignment is useful in a number of bioinformatics applications, such as computing the longest common subsequence of two genes or comparing variants of certain diseases.
[21] Computational neuroscience is the study of brain function in terms of the information processing properties of the nervous system.
[22] Models of the brain include: It is the work of computational neuroscientists to improve the algorithms and data structures currently used to increase the speed of such calculations.
Computational neuropsychiatry is an emerging field that uses mathematical and computer-assisted modeling of brain mechanisms involved in mental disorders.
Several initiatives have demonstrated that computational modeling is an important contribution to understand neuronal circuits that could generate mental functions and dysfunctions.
[28] Analysts project that if major medications fail due to patents, that computational biology will be necessary to replace current drugs on the market.
Doctoral students in computational biology are being encouraged to pursue careers in industry rather than take Post-Doctoral positions.
This is a direct result of major pharmaceutical companies needing more qualified analysts of the large data sets required for producing new drugs.
[28] Computational biology plays a crucial role in discovering signs of new, previously unknown living creatures and in cancer research.
This field involves large-scale measurements of cellular processes, including RNA, DNA, and proteins, which pose significant computational challenges.
Areas of focus include analyzing molecules that are deterministic in causing cancer and understanding how the human genome relates to tumor causation.
Finding centralities in biology can be applied in many different circumstances, some of which are gene regulatory, protein interaction and metabolic networks.
A common supervised learning algorithm is the random forest, which uses numerous decision trees to train a model to classify a dataset.
At each internal node the algorithm checks the dataset for exactly one feature, a specific gene in the previous example, and then branches left or right based on the result.
PLOS cites[citation needed] four main reasons for the use of open source software: There are several large conferences that are concerned with computational biology.