Cutinase
[10] Conversely, the supplementation of cutinase to fungi that are not able to produce it naturally had been shown to enhance fungal infection success rates.
[11] Cutinase belongs to the α-β class of proteins, with a central β-sheet of 5 parallel strands covered by 5 alpha helices on either side of the sheet.
[13] Fungal cutinase is generally composed of around 197 amino acid residues, and its native form consists of a single domain.
[14] The protein also contains 4 invariant cysteine residues that form 2 disulfide bridges, whose cleavage results in a complete loss of enzymatic activity.
[16] This active site is seen flanked by two hydrophobic loop structures and partly covered by 2 thin bridges formed by amino acid side chains.
[15][18] The binding site of the cutin lipid polymer consists of two hydrophobic loops characterized by nonpolar amino acids such as leucine, alanine, isoleucine, and proline.
[18] These hydrophobic residues show a higher degree of flexibility, suggesting an induced fit model to facilitate cutin bonding to the active site.
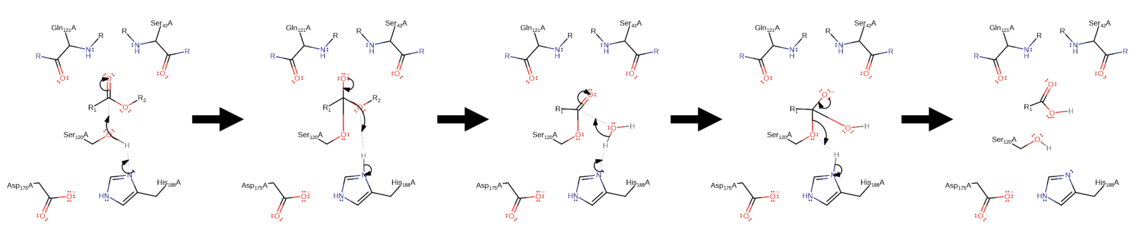
[19] This is followed by an elimination reaction whereby the charged oxygen (stabilized by the oxyanion hole) creates a double bond, removing an R group from the cutin polymer in the form of an alcohol.
[20] In fact, it has been shown that cutinases are more efficient at cleaving and eliminating non-calcium fats from clothing when compared against other industrial lipases.
[21] Another advantage of cutinase in this industry is its ability to be catalytically active with both water- and lipid-soluble ester compounds, making it a more versatile degradative agent.