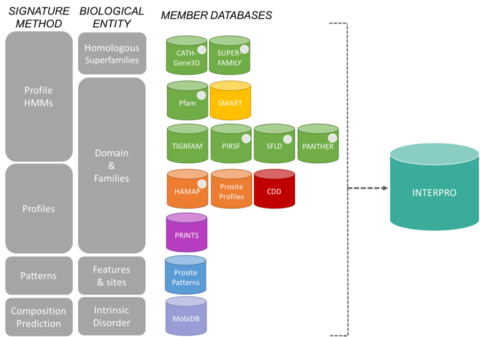
InterPro
InterPro contains three main entities: proteins, signatures (also referred to as "methods" or "models") and entries.
Information regarding which signatures significantly match these proteins are calculated as the sequences are released by UniProtKB and these results are made available to the public (see below).
[5] InterPro also includes data for splice variants and the proteins contained in the UniParc and UniMES databases.
[9] There are six main endpoints for the API corresponding to the different InterPro data types: entry, protein, structure, taxonomy, proteome and set.
InterProScan, along with many other EMBL-EBI bioinformatics tools, can also be accessed programmatically using RESTful and SOAP Web Services APIs.