Mutagenesis (molecular biology technique)
In molecular biology, mutagenesis is an important laboratory technique whereby DNA mutations are deliberately engineered to produce libraries of mutant genes, proteins, strains of bacteria, or other genetically modified organisms.
Mutant strains may also be produced that have practical application or allow the molecular basis of a particular cell function to be investigated.
Since 2013, development of the CRISPR/Cas9 technology, based on a prokaryotic viral defense system, has allowed for the editing or mutagenesis of a genome in vivo.
A number of methods for generating random mutations in specific proteins were later developed to screen for mutants with interesting or improved properties.
[12][13][14] In a European Union law (as 2001/18 directive), this kind of mutagenesis may be used to produce GMOs but the products are exempted from regulation: no labeling, no evaluation.
Other technologies such as cleavage of DNA at specific sites on the chromosome, addition of new nucleotides, and exchanging of base pairs it is now possible to decide where mutations can go.
Such techniques commonly involve using pre-fabricated mutagenic oligonucleotides in a primer extension reaction with DNA polymerase.
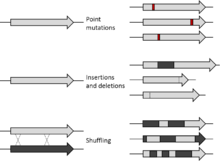
This methods allows for point mutation or deletion or insertion of small stretches of DNA at specific sites.
[25] In this technique, multiple positions or short sequences along a DNA strand may be exhaustively modified to obtain a comprehensive library of mutant proteins.
A segment may also be inserted randomly into the gene in order to assess structural or functional significance of a particular part of a protein.
[27] Engineered mutations such as these can provide important information in cancer research, such as mechanistic insights into the development of the disease.
[28] Transposons, chromosomal segments that can undergo transposition, can be designed and applied to insertional mutagenesis as an instrument for cancer gene discovery.
Our genes occasionally undergo point mutations causing beta-thalassemia that interrupts red blood cell function.
[29] Since 2013, the development of CRISPR-Cas9 technology has allowed for the efficient introduction of different types of mutations into the genome of a wide variety of organisms.
This method allows for extensive mutation at multiple sites, including the complete redesign of the codon usage of a gene to optimise it for a particular organism.