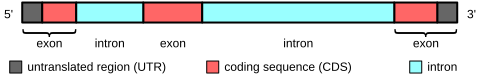
Exon
The term later came to include sequences removed from rRNA[2] and tRNA,[3] and other ncRNA[4] and it also was used later for RNA molecules originating from different parts of the genome that are then ligated by trans-splicing.
[5] Although unicellular eukaryotes such as yeast have either no introns or very few, metazoans and especially vertebrate genomes have a large fraction of non-coding DNA.
[6] This can provide a practical advantage in omics-aided health care (such as precision medicine) because it makes commercialized whole exome sequencing a smaller and less expensive challenge than commercialized whole genome sequencing.
Mature mRNAs originating from the same gene need not include the same exons, since different introns in the pre-mRNA can be removed by the process of alternative splicing.
Splicing can be experimentally modified so that targeted exons are excluded from mature mRNA transcripts by blocking the access of splice-directing small nuclear ribonucleoprotein particles (snRNPs) to pre-mRNA using Morpholino antisense oligos.
However, these sorts of definitions only cover protein-coding genes, and omit those exons that become part of a non-coding RNA[15] or the untranslated region of an mRNA.