Promoter (genetics)
Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand).
Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism.
[citation needed] Promoters represent critical elements that can work in concert with other regulatory regions (enhancers, silencers, boundary elements/insulators) to direct the level of transcription of a given gene.
A promoter is induced in response to changes in abundance or conformation of regulatory proteins in a cell, which enable activating transcription factors to recruit RNA polymerase.
In bacteria, the promoter contains two short sequence elements approximately 10 (Pribnow Box) and 35 nucleotides upstream from the transcription start site.
RNA polymerase holoenzymes containing other sigma factors recognize different core promoter sequences.
Such "closely spaced promoters" have been observed in the DNAs of all life forms, from humans[10] to prokaryotes[11] and are highly conserved.
These events are possible because the RNAP occupies several nucleotides when bound to the DNA, including in transcription start sites.
Eukaryotic RNA-polymerase-II-dependent promoters can contain a TATA box (consensus sequence TATAAA), which is recognized by the general transcription factor TATA-binding protein (TBP); and a B recognition element (BRE), which is recognized by the general transcription factor TFIIB.
An example is the E-box (sequence CACGTG), which binds transcription factors in the basic helix-loop-helix (bHLH) family (e.g. BMAL1-Clock, cMyc).
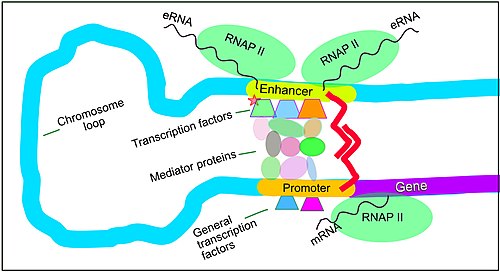
[28] Among this constellation of elements, enhancers and their associated transcription factors have a leading role in the regulation of gene expression.
[30] The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene.
[34] Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two eRNAs as illustrated in the Figure.
Forty-five percent of human somatic oncogenes seem to be regulated by bidirectional promoters – significantly more than non-cancer causing genes.
[43] Certain sequence characteristics have been observed in bidirectional promoters, including a lack of TATA boxes, an abundance of CpG islands, and a symmetry around the midpoint of dominant Cs and As on one side and Gs and Ts on the other.
A motif with the consensus sequence of TCTCGCGAGA, also called the CGCG element, was recently shown to drive PolII-driven bidirectional transcription in CpG islands.
Research suggests that non-coding RNAs are frequently associated with the promoter regions of mRNA-encoding genes.
It has been hypothesized that the recruitment and initiation of RNA polymerase II usually begins bidirectionally, but divergent transcription is halted at a checkpoint later during elongation.
Possible mechanisms behind this regulation include sequences in the promoter region, chromatin modification, and the spatial orientation of the DNA.
[44] The archaeal promoter resembles an eukaryotic one: a TATA box (at -26/-27) and an upstream BRE (at -33/-34) are commonly found, binding to TBP and TFB (homolog of TFIIB).
[3] Strict conservation of these motifs are not necessary, and many archaea with high GC% show "degenerated" TATA boxes.
[7] Another approach is to use a pattern-matching program based on known promoters, from simple hand-crafted regular expressions to advanced machine learning methods such as decision trees, hidden markov models (HMM), and neural networks.
[2] Although RNA polymerase holoenzyme shows high affinity to non-specific sites of the DNA, this characteristic does not allow us to clarify the process of promoter location.
How diseases of different molecular origin respond to treatments is partially addressed in the discipline of pharmacogenomics.
Not listed here are the many kinds of cancers involving aberrant transcriptional regulation owing to creation of chimeric genes through pathological chromosomal translocation.
In humans, DNA methylation occurs at the 5' position of the pyrimidine ring of the cytosine residues within CpG sites to form 5-methylcytosines.
Recent evidence also indicates that several genes (including the proto-oncogene c-myc) have G-quadruplex motifs as potential regulatory signals.
Promoters are important gene regulatory elements used in tuning synthetically designed genetic circuits and metabolic networks.
For example, to overexpress an important gene in a network, to yield higher production of target protein, synthetic biologists design promoters to upregulate its expression.
Automated algorithms can be used to design neutral DNA or insulators that do not trigger gene expression of downstream sequences.
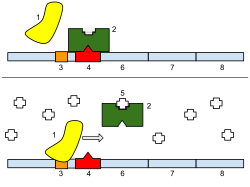
Bottom : The gene is turned on. Lactose is inhibiting the repressor, allowing the RNA polymerase to bind with the promoter and express the genes, which synthesize lactase. Eventually, the lactase will digest all of the lactose, until there is none to bind to the repressor. The repressor will then bind to the operator, stopping the manufacture of lactase.