Eukaryotic translation
Translation initiation is the process by which the ribosome and its associated factors bind to an mRNA and are assembled at the start codon.
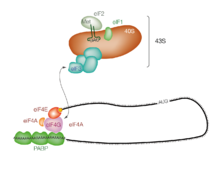
Binding of the cap by eIF4E is often considered the rate-limiting step of cap-dependent initiation, and the concentration of eIF4E is a regulatory nexus of translational control.
Certain viruses cleave a portion of eIF4G that binds eIF4E, thus preventing cap-dependent translation to hijack the host machinery in favor of the viral (cap-independent) messages.
[4] This 43S preinitiation complex (43S PIC) accompanied by the protein factors moves along the mRNA chain toward its 3'-end, in a process known as 'scanning', to reach the start codon (typically AUG).
[citation needed] While protein synthesis is globally regulated by modulating the expression of key initiation factors as well as the number of ribosomes, individual mRNAs can have different translation rates due to the presence of regulatory sequence elements.
In addition, recent work in yeast and humans suggest that evolutionary divergence in cis-regulatory sequences can impact translation regulation.
[6] The best-studied example of cap-independent translation initiation in eukaryotes uses the internal ribosome entry site (IRES).
At the end of the initiation step, the mRNA is positioned so that the next codon can be translated during the elongation stage of protein synthesis.
Eukaryotic mRNA precursors must be processed in the nucleus (e.g., capping, polyadenylation, splicing) in ribosomes before they are exported to the cytoplasm for translation.
Translation can also be affected by ribosomal pausing, which can trigger endonucleolytic attack of the tRNA, a process termed mRNA no-go decay.
Regulation of translation can impact the global rate of protein synthesis which is closely coupled to the metabolic and proliferative state of a cell.
[10] This method enables researchers to take a snapshot of the translatome, showing which parts of the mRNA are being translated into proteins by ribosomes at a given time.
Expanding on this concept, a more recent development is single-cell ribosome profiling, a technique that allows us to study the translation process at the resolution of individual cells.
For instance, activated T cells secrete interferon-γ which triggers intracellular tryptophan shortage by upregulating the indoleamine 2,3-dioxygenase 1 (IDO1) enzyme.