Genomics
A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dimensional structural configuration.
In turn, proteins make up body structures such as organs and tissues as well as control chemical reactions and carry signals between cells.
[13] Following Rosalind Franklin's confirmation of the helical structure of DNA, James D. Watson and Francis Crick's publication of the structure of DNA in 1953 and Fred Sanger's publication of the Amino acid sequence of insulin in 1955, nucleic acid sequencing became a major target of early molecular biologists.
[15][16] Extending this work, Marshall Nirenberg and Philip Leder revealed the triplet nature of the genetic code and were able to determine the sequences of 54 out of 64 codons in their experiments.
[18] Fiers' group expanded on their MS2 coat protein work, determining the complete nucleotide-sequence of bacteriophage MS2-RNA (whose genome encodes just four genes in 3569 base pairs [bp]) and Simian virus 40 in 1976 and 1978, respectively.
[9] In 1975, he and Alan Coulson published a sequencing procedure using DNA polymerase with radiolabelled nucleotides that he called the Plus and Minus technique.
[26][27] For their groundbreaking work in the sequencing of nucleic acids, Gilbert and Sanger shared half the 1980 Nobel Prize in chemistry with Paul Berg (recombinant DNA).
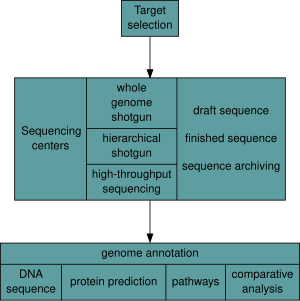
The advent of these technologies resulted in a rapid intensification in the scope and speed of completion of genome sequencing projects.
[33] As of October 2011[update], the complete sequences are available for: 2,719 viruses, 1,115 archaea and bacteria, and 36 eukaryotes, of which about half are fungi.
[34][35] Most of the microorganisms whose genomes have been completely sequenced are problematic pathogens, such as Haemophilus influenzae, which has resulted in a pronounced bias in their phylogenetic distribution compared to the breadth of microbial diversity.
[38][39] The mammals dog (Canis familiaris),[40] brown rat (Rattus norvegicus), mouse (Mus musculus), and chimpanzee (Pan troglodytes) are all important model animals in medical research.
[42] Completion of this project was made possible by the development of dramatically more efficient sequencing technologies and required the commitment of significant bioinformatics resources from a large international collaboration.
[44] The English-language neologism omics informally refers to a field of study in biology ending in -omics, such as genomics, proteomics or metabolomics.
While the growth in the use of the term has led some scientists (Jonathan Eisen, among others[45]) to claim that it has been oversold,[46] it reflects the change in orientation towards the quantitative analysis of complete or near-complete assortment of all the constituents of a system.
However, the Sanger method remains in wide use, primarily for smaller-scale projects and for obtaining especially long contiguous DNA sequence reads (>500 nucleotides).
[60][61] The Illumina dye sequencing method is based on reversible dye-terminators and was developed in 1996 at the Geneva Biomedical Research Institute, by Pascal Mayer and Laurent Farinelli.
To determine the sequence, four types of reversible terminator bases (RT-bases) are added and non-incorporated nucleotides are washed away.
The camera takes images of the fluorescently labeled nucleotides, then the dye along with the terminal 3' blocker is chemically removed from the DNA, allowing the next cycle.
If a homopolymer is present in the template sequence multiple nucleotides will be incorporated in a single flood cycle, and the detected electrical signal will be proportionally higher.
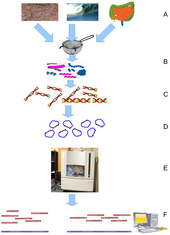
[65][66] Typically the short fragments, called reads, result from shotgun sequencing genomic DNA, or gene transcripts (ESTs).
Other databases (e.g. Ensembl) rely on both curated data sources as well as a range of software tools in their automated genome annotation pipeline.
[76] Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development[77] and tumorigenesis.
[75] The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.
Only very recently has the study of bacteriophage genomes become prominent, thereby enabling researchers to understand the mechanisms underlying phage evolution.
Analysis of bacterial genomes has shown that a substantial amount of microbial DNA consists of prophage sequences and prophage-like elements.
Several studies have demonstrated how these sequences could be used very successfully to infer important ecological and physiological characteristics of marine cyanobacteria.
Thus, the growing body of genome information can also be tapped in a more general way to address global problems by applying a comparative approach.
[86] Genomics has provided applications in many fields, including medicine, biotechnology, anthropology and other social sciences.
[93][94] The All of Us research program aims to collect genome sequence data from 1 million participants to become a critical component of the precision medicine research platform[95] and the UK Biobank initiative has studied more than 500.000 individuals with deep genomic and phenotypic data.
[100] By using genomic data to evaluate the effects of evolutionary processes and to detect patterns in variation throughout a given population, conservationists can formulate plans to aid a given species without as many variables left unknown as those unaddressed by standard genetic approaches.