Genotype
[1] Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location.
In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene.
Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism.
Not all individuals with the same genotype look or act the same way because appearance and behavior are modified by environmental and growing conditions.
Penetrance is the proportion of individuals showing a specified genotype in their phenotype under a given set of environmental conditions.
[7] Traits that are determined exclusively by genotype are typically inherited in a Mendelian pattern.
[8] He studied phenotypes that were easily observed, such as plant height, petal color, or seed shape.
Though Mendel was not aware at the time, each phenotype he studied was controlled by a single gene with two alleles.
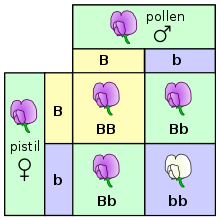
The offspring can inherit a dominant allele from each parent, making them homozygous with a genotype of BB.
Finally, the offspring could inherit a recessive allele from each parent, making them homozygous with a genotype of bb.
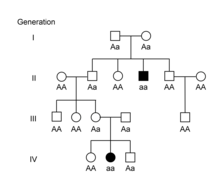
A classic pedigree for an autosomal dominant condition shows affected individuals in every generation.
[13][11][14] In X-linked recessive conditions, males are typically affected more commonly because they are hemizygous, with only one X chromosome.
For example, Huntington disease is an autosomal dominant condition, but up to 25% of individuals with the affected genotype will not develop symptoms until after age 50.
[17] Another factor that can complicate Mendelian inheritance patterns is variable expressivity, in which individuals with the same genotype show different signs or symptoms of disease.
[18][19] For example, a cross between true-breeding red and white Mirabilis jalapa results in pink flowers.
[19] Codominance refers to traits in which both alleles are expressed in the offspring in approximately equal amounts.
[20] A classic example is the ABO blood group system in humans, where both the A and B alleles are expressed when they are present.
The contributions of each of these genes are typically small and add up to a final phenotype with a large amount of variation.
[23] These types of additive effects is also the explanation for the amount of variation in human eye color.
This can be done via a variety of techniques, including allele specific oligonucleotide (ASO) probes or DNA sequencing.
[25] Other techniques are meant to assess a large number of SNPs across the genome, such as SNP arrays.