HMMER
HMMER is a free and commonly used software package for sequence analysis written by Sean Eddy.
[3] HMMER is a console utility ported to every major operating system, including different versions of Linux, Windows, and macOS.
This work is based upon an earlier publication showing a significant acceleration of the Smith-Waterman algorithm for aligning two sequences.
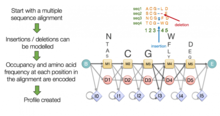
[5] They capitalise on the fact that certain positions in a sequence alignment tend to have biases in which residues are most likely to occur, and are likely to differ in their probability of containing an insertion or a deletion.
Capturing this information gives them a better ability to detect true homologs than traditional BLAST-based approaches, which penalise substitutions, insertions and deletions equally, regardless of where in an alignment they occur.
[7] The HMMER package consists of a collection of programs for performing functions using profile hidden Markov models.
While profile HMM-based homology searches were more accurate than BLAST-based approaches, their slower speed limited their applicability.
[8] The main performance gain is due to a heuristic filter that finds high-scoring un-gapped matches within database sequences to a query profile.
Further gains in performance are due to a log-likelihood model that requires no calibration for estimating E-values, and allows the more accurate forward scores to be used for computing the significance of a homologous sequence.
[12] The major advance in speed was made possible by the development of an approach for calculating the significance of results integrated over a range of possible alignments.