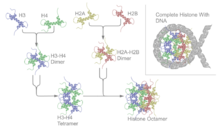
Histone octamer
[2] Richmond and his research group has been able to elucidate the crystal structure of the histone octamer with DNA wrapped up around it at a resolution of 7 Å in 1984.
[3] The structure of the octameric core complex was revisited seven years later and a resolution of 3.1 Å was elucidated for its crystal at a high salt concentration.
[6] Histone tails are subject to a wide array of modifications which includes phosphorylation, acetylation, and methylation of serine, lysine and arginine residues.
Nucleosomes consist of a histone octamer surrounded by 146 base pairs of DNA wrapped in a superhelical manner.
Specific remodeling proteins are constantly altering the chromatin structure by breaking the bonds between the DNA and nucleosome.
The tails play roles both in inter and intra nucleosomal interactions that ultimately influence gene access.
[12] Moreover, these flexible units direct DNA wrapping in a left-handed manner around the histone octamer during nucleosome formation.
Cellular enzymes modify the amino acids in the distal sections of the tail to influence the accessibility of the DNA.
Research has shown removing certain tails prevents the nucleosomes from forming properly and a general failure to produce chromatin fiber.
[12] In all, these associations protect the nucleosomal DNA from the external environment but also lower their accessibility to cellular replication and transcriptional machinery.
No matter the method, in order to modify the nucleosomes, the remodeling complexes require energy from ATP hydrolysis to drive their actions.
In this method, using ATP as an energy source, the translocase domain of the nucleosome-remodeling complex detaches a small region of DNA from the histone octamer.
Once the wave reaches the end of the histone octamer the excess that was once at the edge is extended into the region of linker DNA.
In total, one round of this method moves the histone octamer several base pairs in a particular direction—away from the direction the “wave” propagated.
[6][14] Numerous reports show a link between age-related diseases, birth defects, and several types of cancer with disruption of certain histone post translational modifications.
Studies have identified that N- and C-terminal tails are main targets for acetylation, methylation, ubiquitination and phosphorylation.
[18] Mutations of both p300 have been reported in human tumors such as colorectal, gastric, breast, ovarian, lung, and pancreatic carcinomas.