Nucleic acid analogue
Nucleic acid analogues are compounds which are analogous (structurally similar) to naturally occurring RNA and DNA, used in medicine and in molecular biology research.
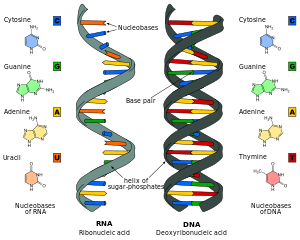
Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pentose sugar, either ribose or deoxyribose, and one of four nucleobases.
Although these oligonucleotides have a different backbone sugar—or, in the case of PNA, an amino acid residue in place of the ribose phosphate—they still bind to RNA or DNA according to Watson and Crick pairing while being immune to nuclease activity.
These nucleoside triphosphates possess a non-canonical sugar, dideoxyribose, which lacks the 3' hydroxyl group normally present in DNA and therefore cannot bond with the next base.
The lack of the 3' hydroxyl group terminates the chain reaction as the DNA polymerases mistake it for a regular deoxyribonucleotide.
The artificial nucleotides featured two fused aromatic rings which formed a (d5SICS–dNaM) complex mimicking the natural (dG–dC) base pair.
[citation needed] Additionally, nitrous acid (HNO2) is a potent mutagen that acts on replicating and non-replicating DNA.
Due to low processivity of the nucleotides linked to bulky adducts such as florophores by [Taq polymerase]s, the sequence is typically copied using a nucleotide with an arm and later coupled with a reactive fluorophore (indirect labelling): Fluorophores find a variety of uses in medicine and biochemistry.
The most commonly used and commercially available fluorescent base analogue, 2-aminopurine (2-AP), has a high-fluorescence quantum yield free in solution (0.68) that is considerably reduced (appr.
[25] Other common tRNA base modifications are pseudouridine (which gives its name to the TΨC loop), dihydrouridine (which does not stack as it is not aromatic), queuosine, wyosine, and so forth.
[33] Introduction of metal ions into a DNA duplex has shown to have potential magnetic[34] or conducting properties,[35] as well as increased stability.
A well-documented example is the formation of T-Hg-T, which involves two deprotonated thymine nucleobases that are brought together by Hg2+ and forms a connected metal-base pair.
These modified nucleobases exhibit tunable electronic properties, sizes, and binding affinities that can be optimized for a specific metal.
Five consecutive copper-hydroxypyridone base pairs were incorporated into a double strand, which were flanked by only one natural nucleobase on both ends.
An unnatural base pair (UBP) is a designed subunit (or nucleobase) of DNA that is created in a laboratory and does not occur in nature.
In 2012, a group of American scientists led by Floyd Romesberg, a chemical biologist at the Scripps Research Institute in San Diego, California, published that his team had designed two unnatural base pairs named d5SICS and dNaM.
[42] More technically, these artificial nucleotides bearing hydrophobic nucleobases feature two fused aromatic rings that form a d5SICS–dNaM complex or base pair in DNA.
[10][43] In 2014, the same team reported that they had synthesized a plasmid containing natural T-A and C-G base pairs along with the best-performing UBP Romesberg's laboratory had designed and inserted it into cells of the common bacterium E. coli, which successfully replicated the unnatural base pairs through multiple generations.
[citation needed] The successful incorporation of a third base pair is a significant breakthrough toward the goal of greatly expanding the number of amino acids which can be encoded by DNA, from the existing 20 amino acids to a theoretically possible 172, thereby expanding the potential for living organisms to produce novel proteins.
[44] Earlier, the artificial strings of DNA did not encode for anything, but scientists speculated they could be designed to manufacture new proteins which could have industrial or pharmaceutical uses.
In 2002, they developed an unnatural base pair between 2-amino-8-(2-thienyl)purine (s) and pyridine-2-one (y) that functions in vitro in transcription and translation, for the site-specific incorporation of non-standard amino acids into proteins.