Off-target genome editing
[1] These tools use different mechanisms to bind a predetermined sequence of DNA (“target”), which they cleave (or "cut"), creating a double-stranded chromosomal break (DSB) that summons the cell's DNA repair mechanisms (non-homologous end joining (NHEJ) and homologous recombination (HR)) and leads to site-specific modifications.
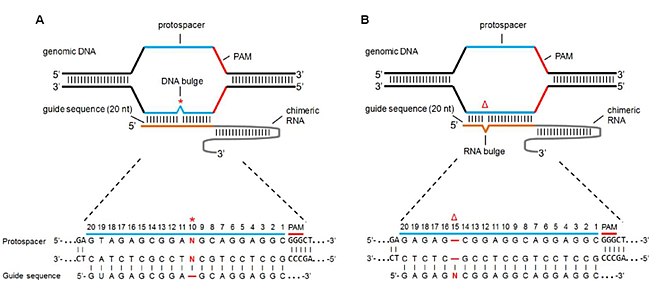
[2] If these complexes do not bind at the target, often a result of homologous sequences and/or mismatch tolerance, they will cleave off-target DSB and cause non-specific genetic modifications.
[12][3] In the research sphere, off-target effects can confound variables in biological studies leading to potentially misleading and non-reproducible results.
[2] In the clinical sphere, the major concerns surround the disruption of vital coding regions leading to genotoxic effects such as cancer.
[13] Accordingly, the improvement of the specificity[14][15] of genome editing tools and the detection[9][16] of off-target effects are rapidly progressing research areas.
Such research incorporates designer nuclease development[17] and discovery,[18] computational prediction programs and databases,[19][20] and high-throughput sequencing[9][16] to reduce and anticipate mutational occurrence.
Many designer nuclease tools are still in their relative infancy and as their molecular properties and in vivo behaviors become better understood they will become increasingly precise and predictable.
[4] Also, the DSBs cleaved by paired nickases have long overhangs instead of blunt ends which provide improved control of targeted insertions.
Additionally, many of the engineering strategies listed above can be combined to create increasingly robust and reliable RNA-guided nuclease editing tools.
Directed evolution can also be used to reduce nuclease activity on particular target sequences, leading to variants such as SpartaCas (containing mutations D23A, T67L, Y128V, and D1251G relative to wildtype SpCas9).
CRISPRi and CRISPRa use a deactivated Cas9 (dCas9) enzyme that cannot cut DNA, but can deliver transcriptional activators and repressors to modulate desired gene expression with high precision.
All of these are listed below: In case of normal targeted sequencing, the biased approach will yield results only for the intended area of capture, which hinders the search as no unexpected mutations will come up on the screen.
[9] The main advantage, however, is that this method is in vitro i.e. the DSBs introduced by Cas9 will not be processed by the DNA repair machinery (unlike BLESS and GUIDE-seq) and thus will include all possible off target mutants.
CHANGE-seq [54] is a more streamlined version of CIRCLE-seq that uses a tagmentation reaction in the circularization step, requiring less DNA material and allowing rapid investigation of off-target sites.
The enrichment of nuclease-induced DNA breaks takes place during PCR amplification, and the protocol is faster than previous in vitro methods with a higher throughput.
BreakTag can map both blunt and staggered DNA breaks, allowing investigation of the different cleavage patterns induced by Cas9 nuclease.
[56] Adapter ligation to quantify cleaved and uncleaved library members allows for unbiased measurement of a nuclease's specificity profile.
Measurement of cleavage of barcoded libraries of targets (BLT) with SpCas9 indicated that specificity profiles were guide-specific and depend on the guide sequence as well as the nuclease itself.
In order for gene editing technologies to make the leap towards safe and widespread use in the clinic, the rate of off-target modification needs to be rendered obsolete.
The safety of gene therapy treatment is of utmost concern, especially during clinical trials when off-target modifications can block the further development of a candidate product.
[57] CAR-T immunotherapy is an ex vivo procedure, which means that the patient's immune cells (in this case T-cells) are extracted and edited using designer nucleases.
[57] While TALEN system development is expensive and time-consuming, research and engineering modifications have drastically limited their rate of off-target interaction.
[58] A phase I/II clinical trial enrolled 12 patients with acquired immune deficiency syndrome (AIDS) to test the safety and effectiveness of administering ZFN-modified autologous helper T cells.
[59] Through targeted deletions, the custom ZFN disables the C-C chemokine receptor 5 (CCR5) gene, which encodes a co-receptor that is used by the HIV virus to enter the cell.
[60] As a result of the high degree of sequence homology between C-C chemokine receptors this ZFN also cleaves CCR2, leading to off-target ~15kb deletions and genomic rearrangements.
By genetically modifying an organism to express an endogenous sequence-specific endonuclease, a target (such as a fertility gene) can be cleaved on the opposite chromosome.
[65] The increased use of genome editing and its eventual translation towards clinical use has evoked controversy surrounding the true off-target burden of the technologies.
On May 30, 2017, a two-page correspondence article was published in Nature Methods that reported an unusually high number of off-target SNVs and indels after sequencing mice that were previously involved in an in vivo gene repair experiment.
[66] The previous experiment, completed by the same group, successfully restored the vision of blind mouse strain (rd1) by correcting the Y347X mutation in the Pde6b gene using a CRISPR-cas9 system.
[70] Nonetheless, off-target rates are consistently found to be more frequent in vivo compared to cell culture experiments, and are thought to be particularly common in humans.